Team:NTNU Trondheim/Characterization
From 2011.igem.org
(→ppGpp's effect on rrnB P1 promoter) |
(→Characterization Results) |
||
| Line 13: | Line 13: | ||
To test the down-regulating effect of ppGpp on rrnB P1, which is the basis of our project, we decided to make a construct containing ppGpp Synthase (RelA) inducible by the pBAD/AraC promoter. The construct would also contain beta-galactosidase (lacZ) which would be expressed by the rrnB P1 promoter. | To test the down-regulating effect of ppGpp on rrnB P1, which is the basis of our project, we decided to make a construct containing ppGpp Synthase (RelA) inducible by the pBAD/AraC promoter. The construct would also contain beta-galactosidase (lacZ) which would be expressed by the rrnB P1 promoter. | ||
Thus, induction of the pBAD/AraC promoter with arabinose should give lower lacZ production, as relA is overproduced giving high ppGpp concentration. This was shown to be the case, using ONPG as a substrate in a lacZ-assay. | Thus, induction of the pBAD/AraC promoter with arabinose should give lower lacZ production, as relA is overproduced giving high ppGpp concentration. This was shown to be the case, using ONPG as a substrate in a lacZ-assay. | ||
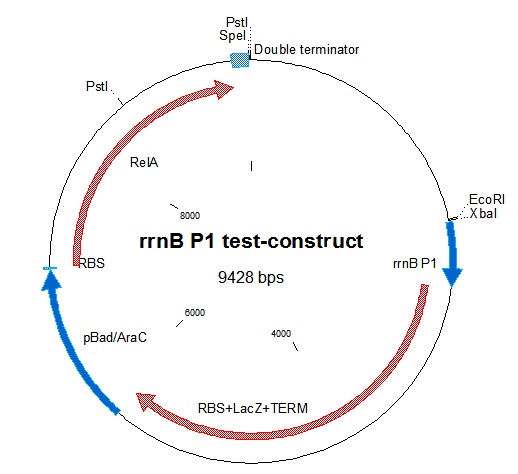
| - | + | [[File:RrnB-relA2.jpg|thumb|Figure 1: Plasmid map of the rrnB P1 test-construct.Note the PstI site in RelA.]] | |
Revision as of 07:16, 24 August 2011

Characterization Results
Several experiments were performed to test our BioBricks and constructs. This page will try to give a short summary of the construction of the test-constructs, the experiments performed to test them, and the results from the experiments.
ppGpp's effect on rrnB P1 promoter
Abstract
To test the down-regulating effect of ppGpp on rrnB P1, which is the basis of our project, we decided to make a construct containing ppGpp Synthase (RelA) inducible by the pBAD/AraC promoter. The construct would also contain beta-galactosidase (lacZ) which would be expressed by the rrnB P1 promoter. Thus, induction of the pBAD/AraC promoter with arabinose should give lower lacZ production, as relA is overproduced giving high ppGpp concentration. This was shown to be the case, using ONPG as a substrate in a lacZ-assay.
Genetic construction
In a previous study, the possible fuction of rrnB P1 in a biological containment system was investigated. ppGpp synthase (relA) was over-expressed to investigate the effect on the promoter. They showed that the expression was completely turned off when high amounts of relA was produced (1). relA synthesizes ppGpp when an "empty" amino-acyl t-RNA binds the ribosome. However when it is over-expressed, ppGpp is produced at sufficient levels to inhibit rrnB P1.
To use relA in our project, we had to amplify the gene from chromosomal DNA, as it was not found in the Partsregistry. Primers were designed as shown below, giving the full 2234 bp gene plus the [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication Openwetware prefix and suffix];
relA.fwd: GTTTCTTCGAATTCGCGGCCGCTTCTAGAGATGGTTGCGGTAAGAAGTGCACA
relA.rev: GTTTCTTCCTGCAGCGGCCGCTACTAGTACTAACTCCCGTGCAACCGACG
The gene was amplified from colony PCR, digested and inserted into digested linearized pSB1A3. After digestion with E+S, it was ligated into E+X-digested [http://partsregistry.org/partsdb/get_part.cgi?part=BBa_B0015 BBa_B0015]. the relA+double terminator was then ligated into an RBS backbone from [http://partsregistry.org/wiki/index.php/Part:BBa_B0034 BBa_B0034]. To be able to control the expression of relA, the arabinose-inducible promoter/inducer [http://partsregistry.org/Part:BBa_K113009 BBa_K113009]containing pBAD/AraC was put in front of the construct.
rrnB P1 was joined with [http://partsregistry.org/Part:BBa_I732019 BBa_I732019]containing an RBS, lacZ coding for beta-galactosidase, and a double terminator. This was then combined with the pBAD-relA construct. A problem we encountered was that relA contains a PstI site at bp 1393. This was mostly worked around in our construction. The final construct is represented in figure 1.
ONPG assay
(1) Tedin, K., A. Witte, et al. (1995). "Evaluation of the E. coli ribosomal rrnB P1 promoter and phage-derived lysis genes for the use in a biological containment system: A concept study." Journal of Biotechnology 39(2): 137-148.
 "
"