Yeast Toolkit Background
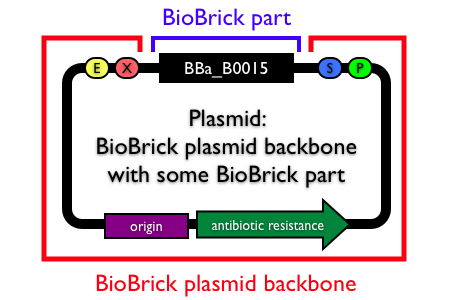
If your iGEM team sets out to do work in E. coli, the Parts Registry provides huge selection of parts and vectors to choose from. But E. coli doesn't make bread. Yeast make bread. If your iGEM team wants to deliver vitamins to malnourished children in a genetically-engineered yeast chassis, there are few resources available to assist you. There is a major gap in the supportive parts - expression cassettes, vectors, and reporters - for iGEM work in yeast.
Finally, with the vitamin A pathway, we are already on our way to a nice reporter library. Beta-carotene is the bright orange pigment found in carrots. Its precursor, lycopene, is the bright red pigment found in ketchup. We decided to go for a third reporter as well: violacein, an unmistakable purple pigment. This will be the first time the violacein pathway will be expressed in a eukaryotic organism. For further details on the purple pigment violacein, please visit the Violacein Page on the Cambridge iGEM 2009 website.
 "
"