Creating the Library of Promoters and UTRs
06/20/11
Purpose: Designing primers for the promoters and UTRs.
07/14/11
Purpose: Do PCR to test operating conditions for Biobrick overhang additions to promoters and UTRs. Run gel on PCR product to test for successful reactions. Teach procedure to Justine, Daphne, Munim and Faraz.
Procedure: James provided me with yeast genomic DNA from Dr. Boeke. We used the primers previously ordered from IDT.
PCR reaction mixture:
- 45µl Platinum Polymerase Supermix
- .6µl Forward Primer
- .6µl Reverse Primer
- 1µl Genomic DNA
- 2.8µl Nuclease Free Water
A general PCR Program of the following form was used:
- 94C for 2mins
- 94C for 30secs-Denaturing
- 48-65C for 30secs-Annealing
- 72C for 1min/kb-Extending
- Repeat the above three steps 30 times.
- 4C-Hold
Round 1:
PCR conducted on:
- ARD1 UTR
- HHO1 Promoter
- PRY1 Promoter
- RPL24A UTR
- STM1 UTR
- TDH3 Promoter
Results:
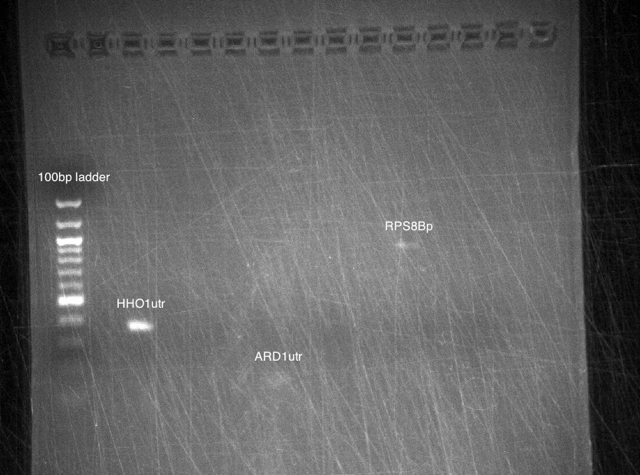
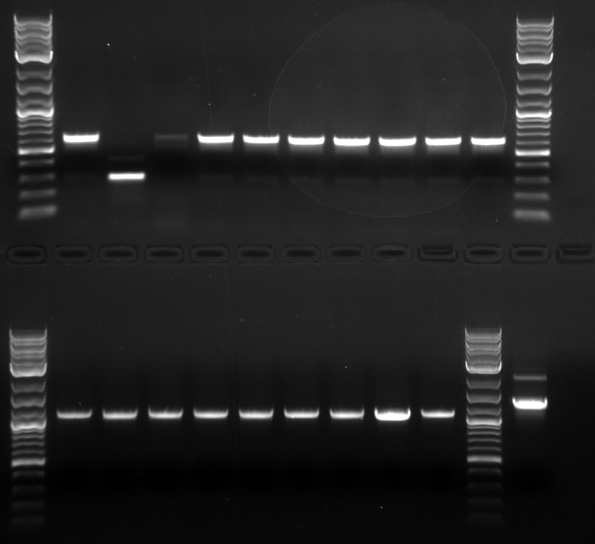
Bands were observed for ARD1 UTR, PRY1 Promoter and STM1 UTR. However, STM1 UTR, with 270bps ran below the band at 250bp. This indicates, that STM1 UTR was a false positive. HHO1 Promoter, RPL24A UTR and TDH3 Promoter show no bands.
PCR conducted on:
- FCY2 Promoter
- Gal1/10 Promoter
- HHo1 Promoter
- RPS8B Promoter
Results: No bands were observed for any of the four promoters. PCR reaction might not have worked due to suboptimal annealing temperature, or self dimerization of the primers.
07/15/11
Purpose: Do PCR to test operating conditions for Biobrick overhang additions to promoters and UTRs. Run PCR on gel to test for successful reaction. Teach procedure to Josh, Sunny and Daniel.
Procedure: See Procedure for 07/13/11.
Round 1:
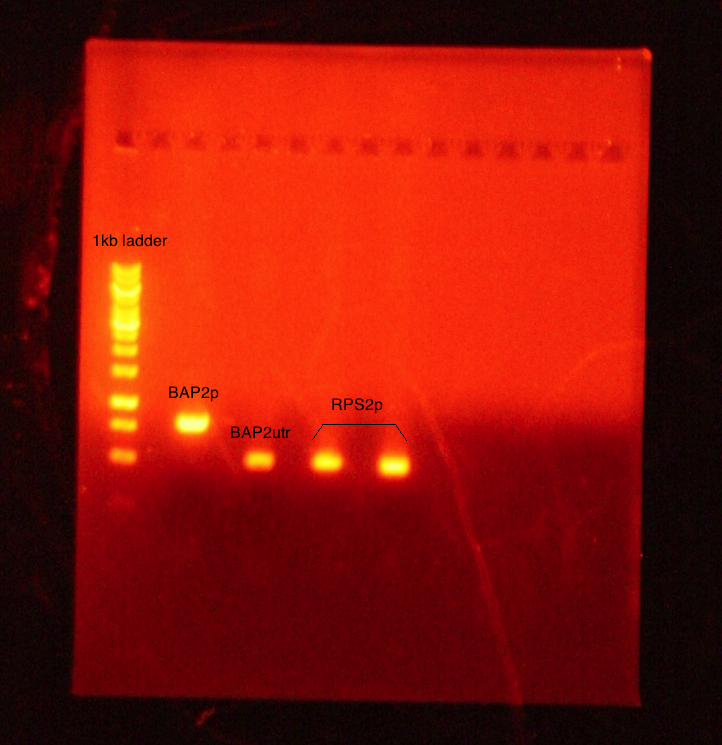
PCR conducted on:
- BAP2 Promoter
- BAP2 UTR
- RPS2 Promoter
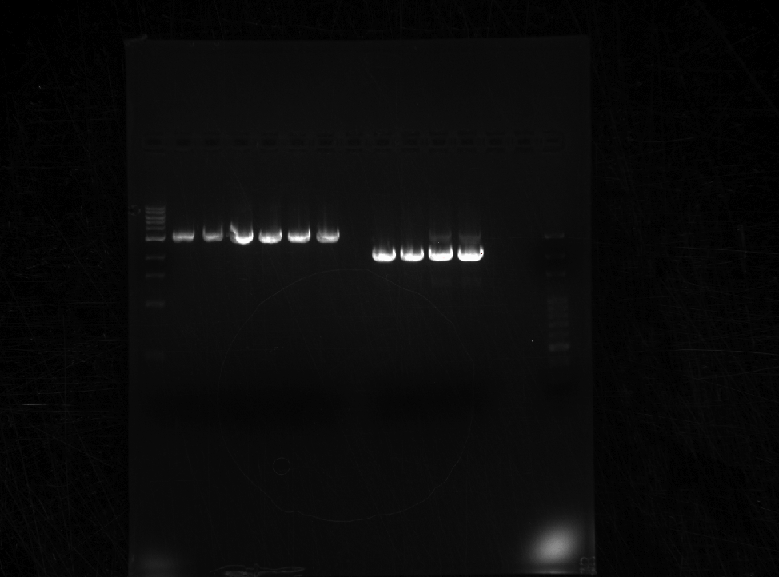
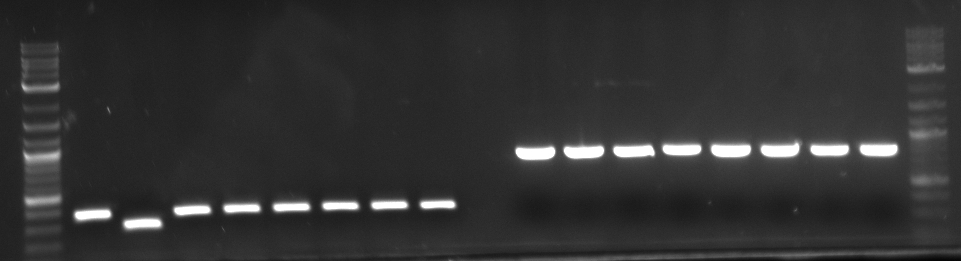
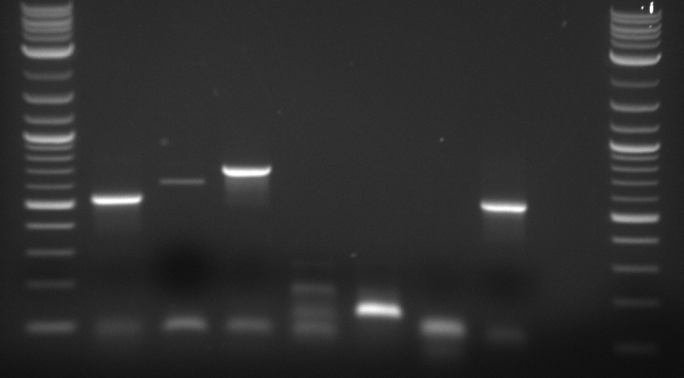
Results: Bands were observed for all three PCR reactions. The fourth band is the same PCR product as the third band. BAP2 Promoter ran above the other three bands at 840bp. The other two products ran to the same position because they are of the same size at 540bp.
Round 2:
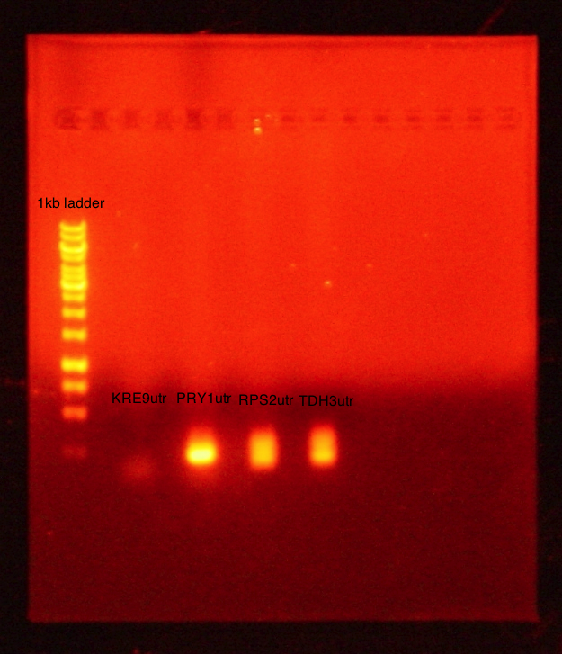
PCR conducted on:
- KRE9 UTR
- PRY1 UTR
- RPS2 UTR
- TDH3 UTR
Results: Bands were observed for three of the four PCR reactions. KRE9 UTR (261bp) did not run a band at all. The other three products ran to the same position because they are of nearly the same size at a range of 140-205bp, as such, the results were inconclusive. We will run another gel with smaller molecular weight ladder on PRY1, RPS2 and TDH3 UTR PCR products to determine whether the PCR worked or not.
07/25/11
Purpose: Run PCRs and gels on the successful promotors and UTRs from 07/14 in order to do gel extraction.
Procedure: See procedure for 07/14/11.
Round 1:
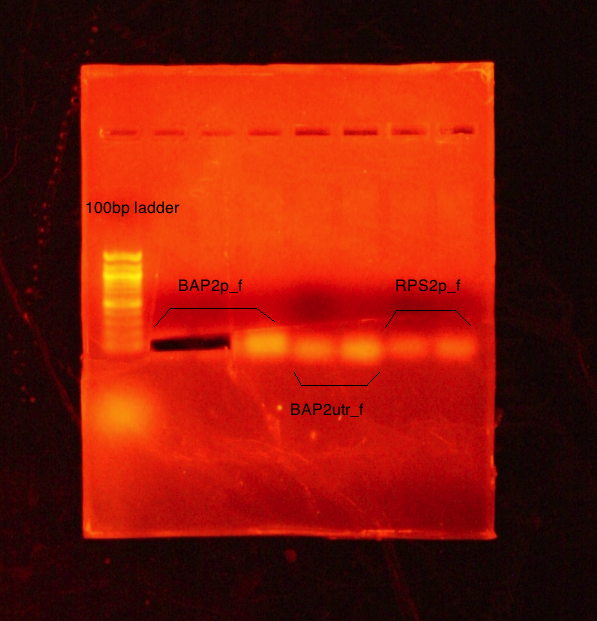
PCR (50µL samples) conducted on:
- BAP2 Promoter
- BAP2 UTR
- RPS2 Promoter
Set annealing temperature to 51C. Ran a 1% agarose gel at 70V.
Results: No desired bands appeared- what appears is too low. PCR reaction might not have worked due to self dimerization of the primers. Bo Jiao (post doc) taught extraction technique using first two wells.
Round 2:
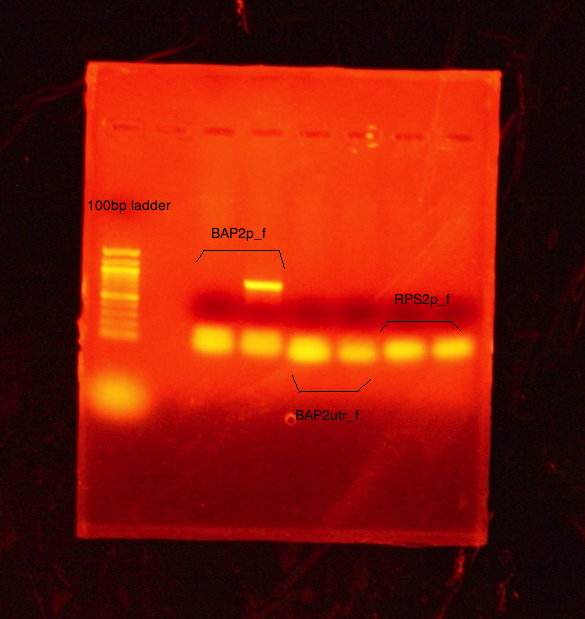
PCR (20µL samples) conducted on:
- BAP2 Promoter
- BAP2 UTR
- RPS2 Promoter
PCR reaction mixture (20µL sample):
- 18µl Platinum Polymerase Supermix
- .24µl Forward Primer
- .24µl Reverse primer
- .4µl Genomic DNA
- 1.12µl Nuclease Free Water
Ran two 20µL samples of each promoter/UTR. Set annealing temperature to 52C. Ran a 1% agarose gel at 90V.
Results A band was observed for the 2nd sample of BAP2p at ~840bp. However, this was decided not enough for extraction. The rest of the samples ran like the previous; bands are inconclusive.
7/28/2011
Purpose: Run gels for analysis of primers and UTRs made on 7/27 and then, based on this analysis, run a gel on the primers and UTRs that are ready for extraction and digestion. Run additional gels for analysis of different primers and UTRs.
Procedure: For PCR and gels, see procedure for 7/14/11.
DNA Digestion Mixture
- 25 μl DNA
- 3 μl Buffer 2
- 1 μl EcoRI-HF
- 1 μl PST1
- 3 μl BSA1
Round 1:
Analysis Gel run on 1 μl samples of:
- TDH3p (wells 2 and 3)
- ARD1utr (3 and 4)
- KRE9p (6)
- STMP1p (7)
- RPL8Autr (8 and 9)
6 μl of 100bp DNA Ladder was used Annealing temperature set to 52C Run at 70V on a 1.5% agarose gel
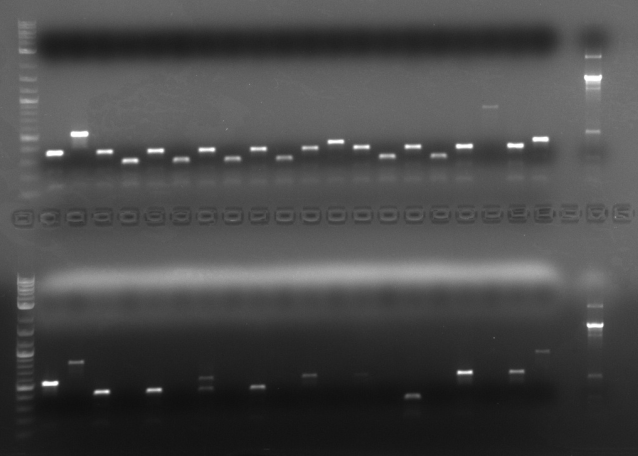
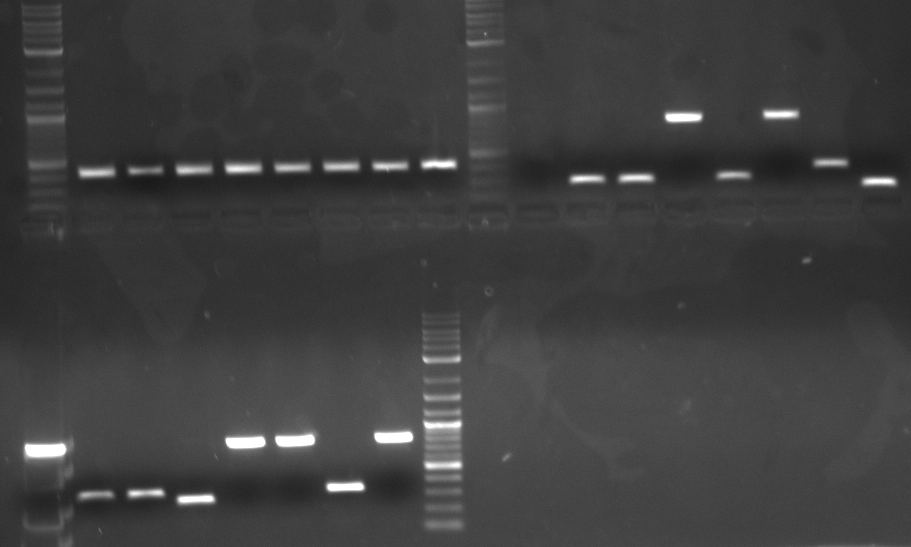
Results The TDH3p (wells 2 and 3), KRE9p (6), and STMP1p (7) all ran to the correct positions and thus were chosen for extraction. The ARD1utr sample wells did not run at all and only one of the two RPL8Autr sample wells ran. A solution, such as changing the temperature for annealing, is being looked into. Extraction and digestion was conducted on TDH3p, KRE9p, and STM1p in addition to the BAP2p and RPS2p extracted on 7/27.
Round 2:
Analysis Gel run on 1 μl samples of:
- HH01utr (well 3)
- FCY2utr (5)
- ARD1utr (7)
- FCY2p (9)
- RPS8Bp (11)
- Gal1/10p (13)
PCR was run at 50C Run on 1.5% gel at 60V 6 μl of 100bp DNA Ladder used
Results The only mixture to show in the gel were HH01utr, ARD1utr, and RPS8Bp suggesting that the annealing temperature needed to be changed.
Round 3:
Analytic Gel run on 1 μl samples of:
- HH01p
- PRY1utr
- KRE9utr
- RPL24Autr
- RPS2utr
- RPS8Butr
- TDH3utr
HH01p, PRY1utr, and KRE9utr were run in PCR at an annealing temperature of 52C RPL24Autr, RPS2utr, RPS8Butr, and TDH3utr were run in PCR at an annealing temperature of 48C Run on a 1.5% gel at 60V 6 μl of 100bp DNA Ladder used
Results: None of the mixes ran in the gel due to the use of expired PCR Supermix.
7/29/2011
Purpose: Run an analytic gel for the promoters and UTRs made on 7/28 with intent to extract. Try to amplify more promoters and UTRs.
Procedure:
ROUND 1
We ran a 1.5% agarose gel @70V for the following (PCR'd at annealing temperature 51C):
- HH01p
- PRY1p
- FCY2p
- PRY1utr
- KRE9utr
Results:
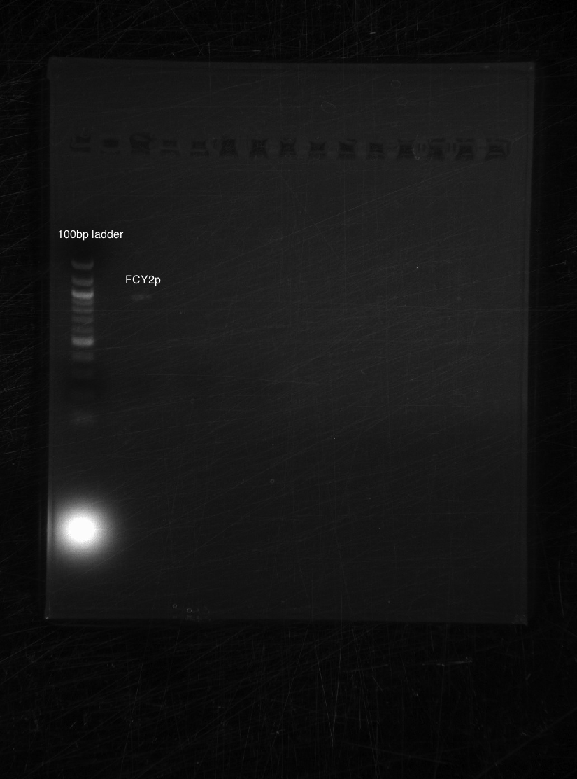
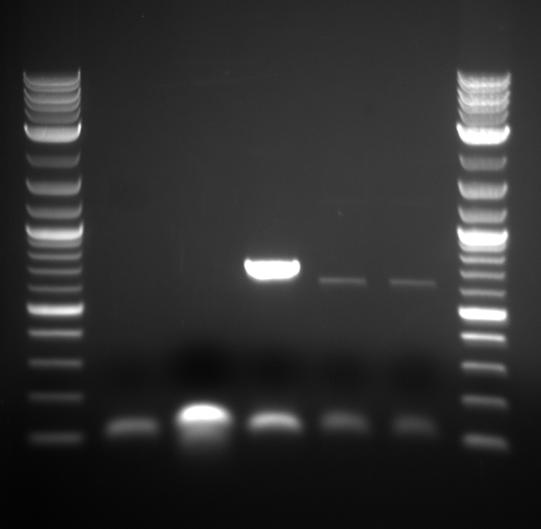
Bands appeared at the correct positions for HH01p, PRY1p, FCY2p, and KRE9utr. Although bright (save for HH01p), they were determined not enough for extraction because of size vs. quantity; more was made of each to extract at a later date. PRY1utr had no observable bands.
ROUND 2
We ran a 1.5% agarose gel @60V for the following:
- FCY2p
- FCY2utr
- Gal 1/10p
Results:
As expected, only FCY2p worked. We're still trying to optimize conditions for FCY2utr and Gal 1/10p.
8/01/2011
Purpose: Run an analytic gel for the promoters and UTRs made on 7/29. Extract successful P/U. Try to amplify more.
Procedure:
ROUND 1
We ran a 1.5% agarose gel @70V for the following (PCR'd at annealing temperature 48C):
- ARD1utr
- RPL24Autr
- RPS8utr
- TDH3utr
- RPS2utr
Results:
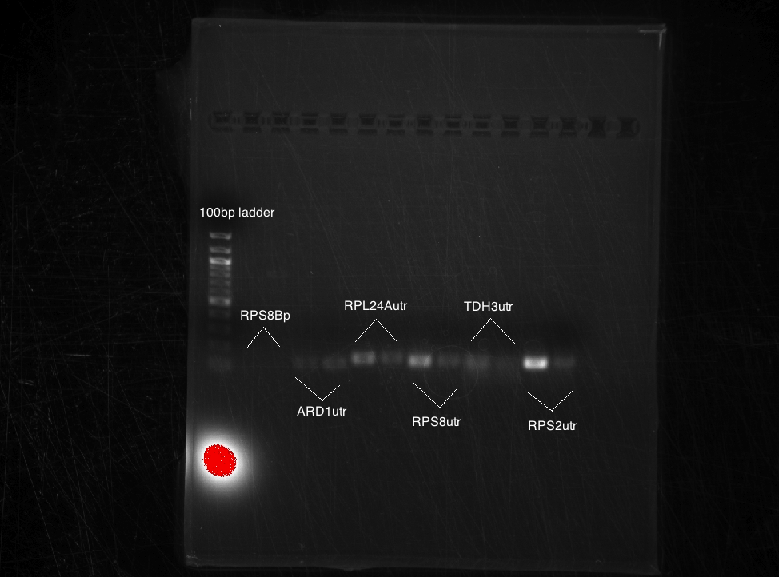
Unfortunately, no bands appeared. Suggested annealing temperature might be too low.
ROUND 2
We made more FCY2p, KRE9utr, PRY1p, HH01utr to use for extraction (protocol on wiki). However, we chose not to extract FCY2p yet because since it's bigger (length), more would be needed for a higher DNA concentration. ~40µl extracted: 550mg KRE9utr, 310mg PRY1p, 570mg HH01utr. 1 gel volume of isopropanol was added to KRE9utr and HH01utr since protocol suggests for x<500bp to increase DNA concentration. Digested three samples using same procedure as other digestions. Stored at -20C.
8/02/2011
Purpose: Increase concentrations of selected promoters and UTRs from 5µM to 10µM. Run through gel to find optimal concentration.
Procedure:
ROUND 1
We ran a 1.5% agarose gel @70V for the following (PCR'd at annealing temperature 53C):
- ARD1utr 5µM
- ARD1utr 10µM
- RPL24Autr 5µM
- RPL24Autr 10µM
- RPS8utr 5µM
- RPS8utr 10µM
- TDH3utr 5µM
- TDH3utr 10µM
- RPS2utr 5µM
- RPS2utr 10µM
- RPS8Bp 5µM
- RPS8Bp 10µM
8/03/2011
Purpose: iGEM vector and backbones arrived- need to be resuspended and PCR'd.
Procedure:
ROUND 1
Resuspended BBAP to 60µM. Below is the backbone PCR (20µL) protocol using Phusion:
- 4µl 5x Phusion HF buffer
- 0.4µl 10mM dNTPs
- 2µl primers diluted to 0.5µM
- 0.5µl template DNA
- 0.2µl Phusion DNA polymerase
- 12.9µl H2O
PCR Program used:
- 98C for 30secs
- 98C for 10secs-Denaturing
- 53C for 30secs-Annealing
- 72C for 90secs-Extending
- 72C for 8min-Final Extension
- Repeat the above three steps 30 times.
- 4C-Hold
We ran a 1.5% agarose gel @70V for the following:
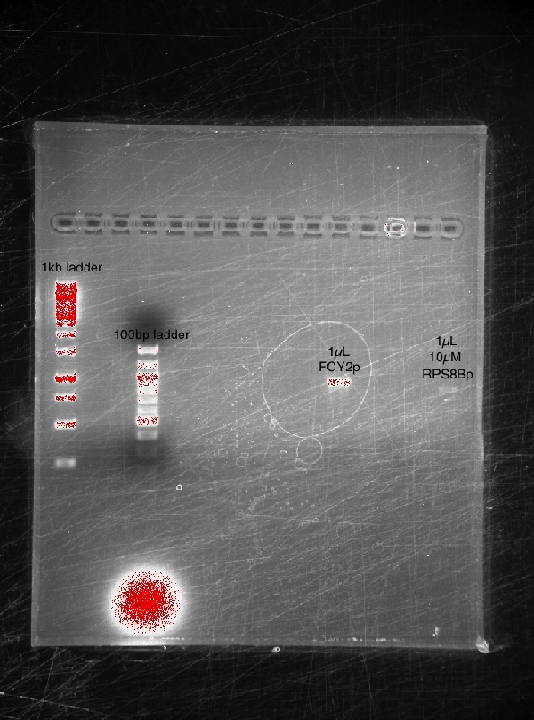
- BBAP (PCR'd @ 53C)
- FCY2p - 53C
- RPS8Bp 5µM - 53C
- RPS8Bp 10µM - 53C
We ran a 1.5% agarose gel @70V for the following (PCR'd at annealing temperature 53C):
- RPL24Autr 5µM
- RPS8Butr 5µM
- RPS2utr 5µM
ROUND 3
Ran 4 extraction wells for FCY2p made on 07/28 (51C), 07/29, 08/01 (51C), 08/02 (53C) ~450mg. Digested sample using same procedure as other digestions. Stored at -20C.
8/04/2011
Purpose: Get iGEM vectors and backbone to work. Run an analytic gel for the BBAP(08/03) and P/U(08/02). Try to amplify more. Extract BBAP if successful.
Procedure:
ROUND 1
We ran a 1.5% agarose gel @60V for the following:
- BBAP (PCR'd @ annealtemp 55C)
- RPL24Autr
- RPS8Butr
- RPS2utr
ROUND 2
Made 4 more tubes of BBAP with annealtemp 55C. Ran 6 extraction wells for BBAP: 1st sample: 340mg, 2nd sample: 300mg, 3rd sample: 300mg. Digested samples:
- 30µl DNA
- 3.5µl buffer 2
- 0.75µl BSA
- 0.75µl PST1
- 0.75µl EcoRI-HF
(Stored at -20C)
8/05/2011
Purpose: Try to amplify more promoters and UTRs. With successful BBa, try to ligate/transform with BAP2p and BAP2utr.
Procedure:
ROUND 1We ran a 1.5% agarose gel @55V for the following (PCR'd @ annealtemp 51C):
- RPS8Bp
- HHO1p
- ARD1utr
- RPL8Autr
- TDH3utr
ROUND 2
First found concentrations of BBa, BAP2p, and BAP2utr. After vacuufuging, concentrations are as follows: BBa 13.2ng/µl, BAP2p 10.3ng/µl, BAP2utr 12.9ng/µl. Ligation procedure for BBa + BAP2p:
- 3.84µl BAP2p
- 1µl BBa
- 2µl ligase buffer
- 2.16µl H2O
- 1µl T4 ligase
Ligation procedure for BBa + BAP2utr:
- 3.06µl BAP2utr
- 1µl BBa
- 2µl ligase buffer
- 2.94µl H2O
- 1µl T4 ligase
Inoculated in 10mL LB agar with 19µl 34mg/mL chloromphenicol. FORGOT to add agar to ligations after shaking in incubator. 16 hours later, BBa plates grew colonies but the two ligations did not.
8/11/2011
Purpose: Amplify P/U for ligation/transformation.
Procedure: We ran a 1.5% agarose gel @70V for the following (PCR'd @ annealtemp 52C):
- TDH3p
- STM1p
- RPL8Autr
Then, we did PCR purification and digestion on the samples.
8/12/2011
Purpose: Amplify P/U for ligation/transformation.
Procedure: We ran a 1.5% agarose gel @70V for the following (PCR'd @ annealtemp 51C):
- PRY1p
- FCY2p
- KRE9utr
Then, we did PCR purification and digestion on the samples.
8/15/2011
Purpose: Amplify more promoters and UTRs. With successes, ligate/transform with backbone.
Procedure:
ROUND 1
We ran a 1.5% agarose gel @70V for the following (PCR'd @ annealtemp 50C):
- HHO1utr 5µM
- ARD1utr 10µM
- RPS8Bp 10µM
Results:
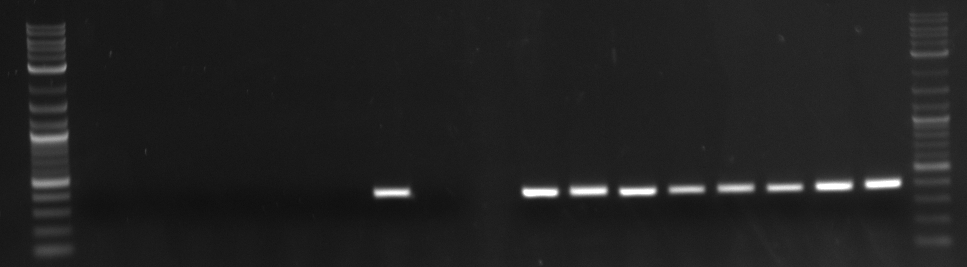
Sufficient bands showed for only HHO1utr and RPS8Bp.
ROUND 2
Dephosphorylated backbone with CIP:
- 26µl DNA
- 3µl buffer 2
- 1µl CIP
Ran through an extraction gel.
8/16/2011
Purpose: Try to create first BioBrick.
ROUND 1Procedure: Miniprepped BAP2p and did a single-cut digestion:
- 21µl DNA
- 2.5µl buffer 4
- 0.25µl BSA
- 0.83µl Xba1
- 0.42µl H2O
Ran through analytic gel.
ROUND 2
Found concentrations of P/U thus far after another PCR clean and vacuufuging.
| Component | Concentration (ng/µl) |
| RPL8Autr | 5.6 |
| STM1p | 20.4 |
| KRE9utr | 11.7 |
| PRY1p | 18.4 |
| FCY2p | 49.2 |
| HHO1utr | 15.1 |
| TDH3p | 9.8 |
| RPS8Bp | 14.1 |
| BBa | 61.3 |
Ligation procedure for BBa + FCY2p:
- 0.70µl FCY2p
- 1.23µl BBa
- 2µl ligase buffer
- 5.07µl H2O
- 1µl T4 li
08/24/11
Confirming the validity of the psb1c3 biobrick. In order to verify that the DNA we have is actually the psb1c3 backbone with the RFP part, I digested with XbaI and EcorI and ran uncut plasmid beside it to assess how it runs when uncut.
08/26/11
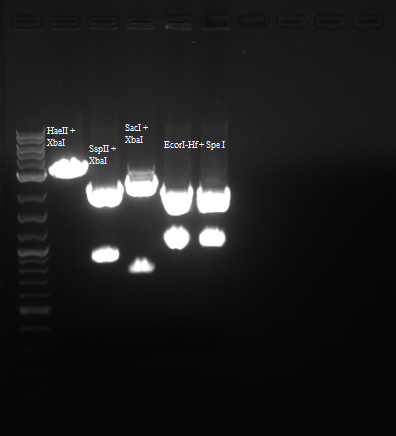
In order to confirm that we do indeed have our backbone with the RFP part in it I digested with the enzymes below:
| Enzyme 1 | Enzyme 2 | Buffer | BSA Added? |
| EcorI-Hf | SpeI | 4 | Yes |
| SSpII | XbaI | 4 | Yes |
| HaeII | XbaI | 4 | Yes |
| SacI | XbaI | 4 | Yes |
Results were as follows:
Also, I proceeded to PCR amplify our inserts from the gDNA of yeast.
| Volume uL | |
| Herculase 5X Buffer | 10 |
| 2.5mM dNTP Mix | 5 |
| 100-300ng genomic DNA | X |
| 10uM Forward Primer | 1.25 |
| 10uM Reverse Primer | 1.25 |
| Herculase II Enzyme | 1 |
| Sterile Water | Y |
| Total | 50 |
| Temperature | Time | Cycles |
| 95 Celsius | 2 minutes | 1 |
| 95 Celsius | 20 seconds | 30 |
| 55 Celsius | 20 seconds | 30 |
| 72 Celsius | 30 seconds | 30 |
| 72 Celsius | 3 minutes | 1 |
08/28/11
After PCR purification of my inserts I proceeded to digest the inserts and a psb1c3 vector with the following mixture:
| Reagent | Volume uL |
| NEB Buffer 4 | 3 |
| Insert | 25 |
| EcorI | 1 |
| SpeI | 1 |
| BSA | .3 |
The digestion mixture was then heat inactivated at 60 Celsius for 20 minutes. All 30uL were then loaded on a gel and gel extracted to ensure that the ssDNA removed by digestion doesn't religate. The gel piece was then gel purified through the use of Qiagen Gel Extraction Kit and eluted with 20uL Tris HCl at pH7.4.
I then proceeded to ligate the inserts with the E+S digested backbone.
| Reagent | Volume uL |
| 10X T4 Lig. Buffer | 1.5 |
| Insert | 6 |
| Vector | 3 |
| T4 Ligase | 1 |
| sterile water | 3.5 |
Ligation was set to run overnight at 16 Celsius.
08/30/11
I proceeded to transform my ligated product into E.coli with a heat shock of 45 seconds. I grew the transformants up for an hour in SOC media. Finally, the transformants were plated into chloroamphenicol plates.
08/31/11
The positive control RFP plate showed colonies as normal, however, all the other parts showed no colonies whatsoever. Will need to find mistake.
09/01/11
Performed PCR on my inserts once more.
| Herculase 5X Buffer | 10 |
| 2.5mM dNTP Mix | 5 |
| 100-300ng genomic DNA | X |
| 10uM Forward Primer | 1.25 |
| 10uM Reverse Primer | 1.25 |
| Herculase II Enzyme | 1 |
| Sterile Water | Y |
| Total | 50 |
| Temperature | Time | Cycles |
| 95 Celsius | 2 minutes | 1 |
| 95 Celsius | 20 seconds | 30 |
| 55 Celsius | 20 seconds | 30 |
| 72 Celsius | 30 seconds | 30 |
| 72 Celsius | 3 minutes | 1 |
Proceeded to do PCR clean and digest the pieces with EcorI and SpeI.
09/02/11
Did gel extraction in order to separate the inserts from the small ends ssDNA removed through digestion.
Set up a ligation of the 18 inserts overnight.
09/02/11
Transformed the ligation product into 50ul competent DH5alpha cells.
09/03/11
16 plates out of the 18 had plenty of colonies. Negative control had no colonies and RFP positive control had colonies.
Picked 10 colonies per plate and grew them up in LB media with 34ug/mL chloroamphenicol.
09/04/11
Stored samples in 4 Celsius cold room.
09/06/11
Performed csPCR on sample of colonies. To do so, first added 4ul of innoculated culture to 50uL of water and set it at 95 Celsius for 10 minutes. Then, took 2uL of this mixture and added it as template to the csPCR mixture. First round of csPCR was done only on two inserts.
| Reagents | Volume uL |
| Buffer | 10 |
| 2.5mM dNTP Mix | 5 |
| 100-300ng genomic DNA | X |
| 10uM F. Sequencing Primer | 1.25 |
| 10uM R. Sequencing Primer | 1.25 |
| rTaq Polymerase | 1.25 |
| Sterile Water | Y |
| Total | 25 |
| Temperature | Time | Cycles |
| 95 Celsius | 2 minutes | 1 |
| 95 Celsius | 20 seconds | 25 |
| 55 Celsius | 20 seconds | 25 |
| 72 Celsius | 30 seconds | 25 |
| 72 Celsius | 3 minutes | 1 |
Sequencing primers were designed such that they have 150bp regions flanking the biobrick on either side. This allows for better sequencing results later on, however, this also adds 300bp on the total gene size when running csPCR.
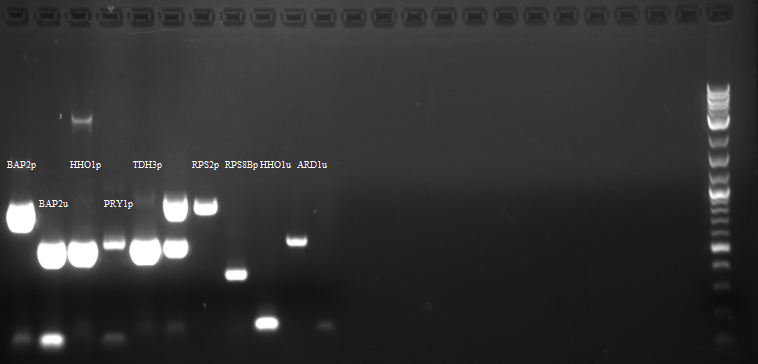
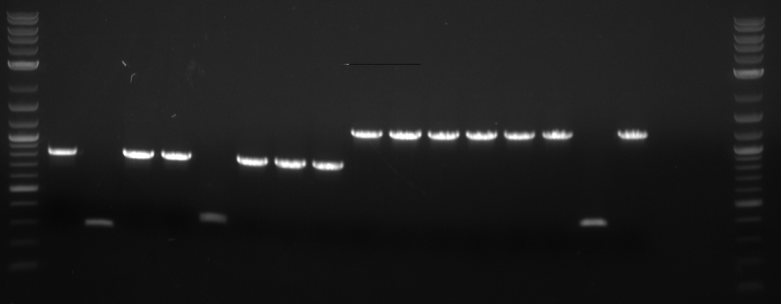
Ran the PCR products of HHO1u and BAP2p on a gel.
Both were of correct size.
09/07/11
Repeated csPCR procedure on 6 other sets of cultured inserts in the cold room.
Performed zymogen miniprep protocol on HHO1u and BAP2p.
Prepared to send for sequencing by adding 500ng in 15uL water with 5uM primer, prepared individual tubes for forward and backward read.
09/08/11
Attempted to PCR 6 of the inserts that I had not yet attempted to extract.
Repeated csPCR on 7 more inserts.
Prepared confirmed inserts from yesterday's csPCR for sequencing.
Picked more colonies from plates of FCY2p and RPS2p because of csPCR failure.
09/09/11
Performed csPCR on FCY2p and RPS2p colonies.
Sent 16 inserts for sequencing.
09/10/11
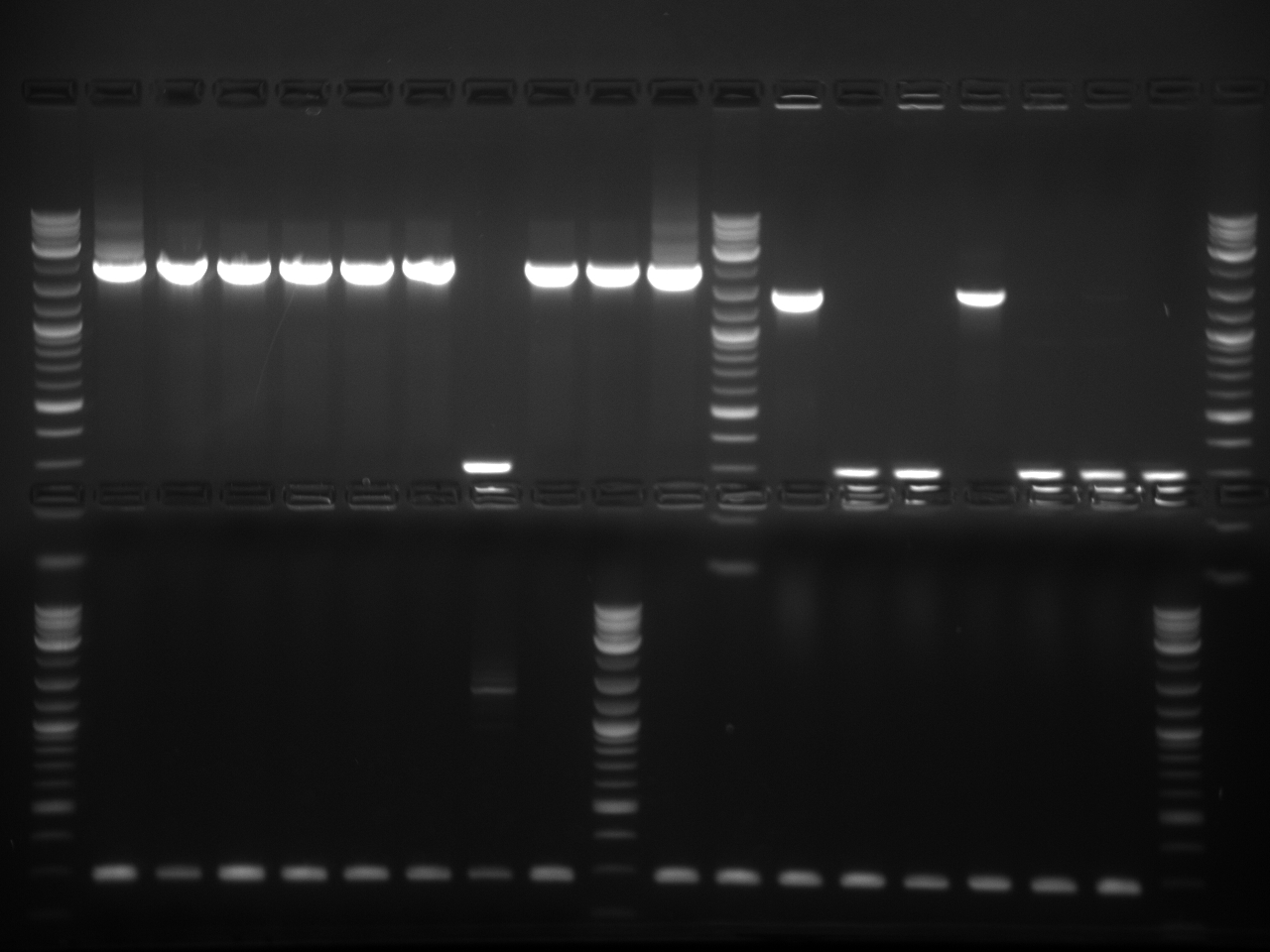
Received sequences from Genewiz. After analyzing the trace files with APE, it seems that I had 10 correct inserts to send as biobrick parts.
Performed standard PCR on Vitamin A genomic strains in order to determine the expression of each strain and whether the strains were what we thought or not.
Results were inconclusive. However, the gel did determine that our sequences at their correct sizes were indeed within these strains.
09/11/11
Prepped another set of samples for sequencing.
09/12/11
Ran PCR on the inserts that had failed previously at a higher melting temperature.
Inserts 1, 3 and 5 showed bands at correct size.
Sent samples to genewiz for sequencing.
Used PCR to extract our vitamin A genes from the genome of our strains. PCR purified these selfsame inserts and digested. Proceeded to ligate overnight.
09/13/11
Received sequencing information from genewiz. Three more sequences were confirmed.
Transformed our vitamin A genes ligated to pcb1c3 into e.coli.
Picked colonies from successful transformation to grow overnight.
09/14/11
Performed csPCR on vitamin A genes.
CrtI and E showed bands of the correct size. However, by mistake I discarded the culture of overnight E.coli for CrtE.
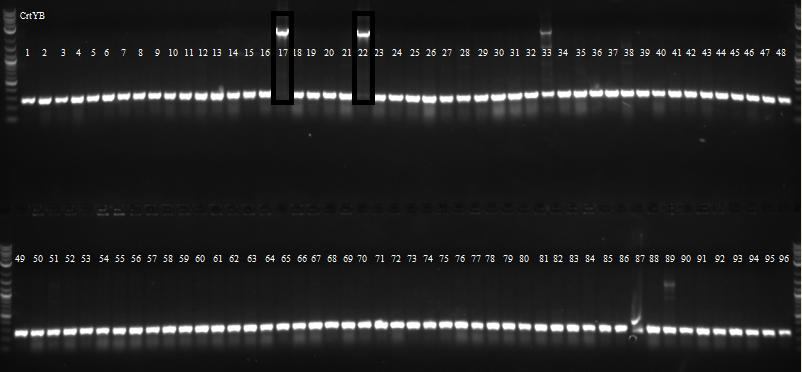
Picked 96 colonies from both CrtE and CrtYB for overnight growth in 96 well plates for csPCR.
09/15/11
csPCR of CrtYB showed that only two colonies had bands of the correct size.
csPCR of CrtE showed many colonies had bands of the correct size.
Sent 4 CrtI, 4 CrtE and 2 CrtYb colonies for sequencing.
09/16/11
Sequencing results returned from genewiz. Both CrtE and CrtI had colonies with the correct sequence, however, CrtYB only had religated vector, as the chloroamphenicol gene was the one sequenced.
09/20/11
Prepared csPCR of second round of vitaminA clones and qcPCR clones.
| Parts | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| CrtI | + | + | + | + | + | + | + | - | - | - | - |
| CrtE | + | + | + | + | + | + | + | + | - | - | - |
| CrtYB | + | + | + | + | + | + | + | + | + | - | - |
| Gal1/10 | + | + | + | - | - | - | - | - | - | - | - |
| Empty | - | - | - | - | - | - | - | - | - | - | - |
| Empty | - | - | - | - | - | - | - | - | - | - | - |
| CrtI Pc | + | + | + | + | + | + | + | + | + | - | - |
| CrtE Nc | + | + | + | + | + | + | + | + | + | - | - |
Ran in PCR machine.
| Temperature | Time | Cycles |
| 95 Celsius | 2 minutes | 1 |
| 95 Celsius | 20 seconds | 20 |
| 56 Celsius | 20 seconds | 20 |
| 72 Celsius | 30 seconds | 20 |
| 72 Celsius | 3 minutes | 1 |
Grew RPL8Au and RPS8Bu in LB chloro overnight.
Transformed Gal1 promoter from registry into cells.
Characterizing the Promoters and UTRs
Measuring the Activity of BioBrick Promoters Using an In Vivo Reference Standard (published in 2009).
9/9/2011
Reference standards received from Imperial College London
Plan: To clone the vectors in E. coli using the Amp selection marker, then miniprep, cut at StuI and transform into yeast.
9/10/11
Standards from Imperial College London transformed into E. coli (protocol specified in the Protocol section).
Cells with plasmids with promoters are plated on LB+Carb plate, and incubated at 37oC overnight.
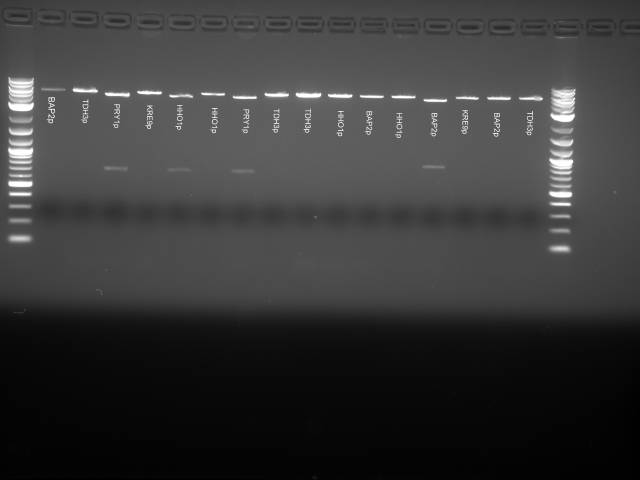
About 50µL of liquid culture taken from Daniel Wolozny for following inserts:
- Bap2p
- TDH3p
- PRY1p
- KRE9p
- HHO1P
- STM1P
9/11/11
Plates incubated on the previous day are taken out. Colonies grew on the plates for the reference standards. Lawn growth resulted on the the plates for the inserts.
Decide to keep on with the Lawn growth plates anyway; the cultures used to grow the plates were grown from a single colony, so all the cells should be identical.
Take a colony from each plate (or a small section of the plate) into 5 mL of LB+Carb.
9/12/11
Miniprep the grown cultures.
The grown cultures were miniprepped via Zyppy Plasmid Miniprep protocol (Zymo Research).
Concentrations was determined by nanodrop.
9/14/11
Digest by EcoRI and SpeI enzyme, with protocol as specified in the protocol page in the BioBrick protocol.
The provided plasmids from Imperial College London with the reference promoters were digested with StuI enzyme.
Digestions done at 37oC for one hour.
9/16/11
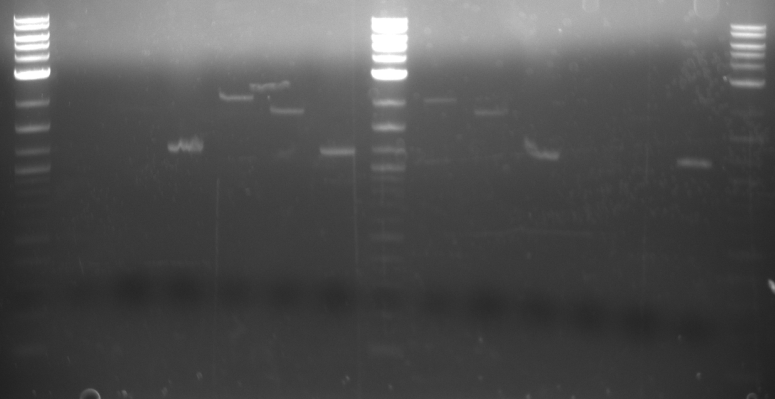
A gel was ran on the digested products to check for products. Products confirmed. One of the three plasmids provided by Imperial College London showed complete digestion.
Proceed to Gel extraction using Qiagen gel extraction kit.
9/18/11
Yeast (YPH500) inoculated for transformation
9/19/11
Yeast transformation:
the StuI digested reference vectors are transformed into the YPH500 culture via protocol specified in the protocol page.
Plated at two concentration of 50 and 350 µL per plate.
Ligation also done for the inserts into the cyc1p EcoRI and SpeI digested plasmids (specific calculation will be included once
9/23/11
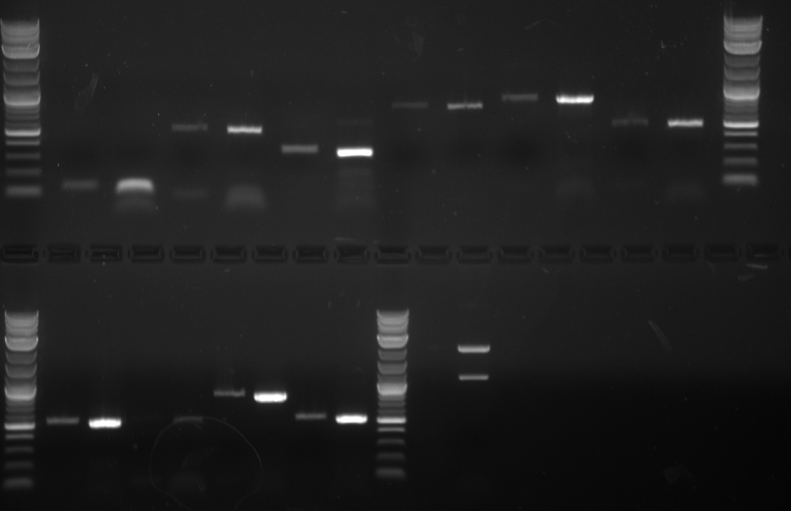
(Second digestion with EcoRI and SpeI are done by Daniel Wolozny and Omar Hadzipasic) A gel is done on the digested samples for verification
This gel shows odd results. The only one with the right size band is the BAP2p plasmid. All the bands in the gel seemed linearized, so it is possible that the plasmids were only cut once.
We decided to run a PCR with the primers for the inserts to check if the inserts were present.
PCR with GoTaq Green (Promega):
| Ingredient | Volume (µL) for each one reaction |
| GoTaq Mix | 12.5 |
| Primer mix (10nM) | 2.5 |
| DNA Template | 2 |
| Nuclease Free water | 8 |
 "
"