|
|
| (10 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{:Team:Johns_Hopkins/Templates/tpl1}} | | {{:Team:Johns_Hopkins/Templates/tpl1}} |
| - | <html> | + | <html><br/> |
| - | <div id="secondarycontentgreen"> | + | <center><table id="Table_01" width="664" height="35" border="0" cellpadding="0" cellspacing="0"> |
| | + | <tr> |
| | + | <td> |
| | + | <a href="https://2011.igem.org/Team:Johns_Hopkins/Notebook/VitExperiments"><img src="https://static.igem.org/mediawiki/2011/3/38/Menu2vit_01.jpg" width="195" height="35" alt=""></a></td> |
| | + | <td> |
| | + | <img src="https://static.igem.org/mediawiki/2011/3/37/Menu2vitblack_02.jpg" width="30" height="35" alt=""></td> |
| | + | <td> |
| | + | <a href="https://2011.igem.org/Team:Johns_Hopkins/Notebook/BCAssay"><img src="https://static.igem.org/mediawiki/2011/3/34/Menu2vitblack_03.jpg" width="177" height="35" alt=""></a></td> |
| | + | <td> |
| | + | <img src="https://static.igem.org/mediawiki/2011/b/bc/Menu2vitblack_04.jpg" width="29" height="35" alt=""></td> |
| | + | <td> |
| | + | <a href="https://2011.igem.org/Team:Johns_Hopkins/Notebook/DoughMed"><img src="https://static.igem.org/mediawiki/2011/e/e8/Menu2vitblack_05.jpg" width="120" height="35" alt=""></a></td> |
| | + | <td> |
| | + | <img src="https://static.igem.org/mediawiki/2011/d/d0/Menu2vitblack_06.jpg" width="30" height="35" alt=""></td> |
| | + | <td> |
| | + | <a href="https://2011.igem.org/Team:Johns_Hopkins/Notebook/Baking"><img src="https://static.igem.org/mediawiki/2011/7/72/Menu2vitblack_07.jpg" width="83" height="35" alt=""></a></td> |
| | + | </tr> |
| | + | </table></center> |
| | + | <div id="secondarycontentgreen"> |
| | <div id="boxheading"> | | <div id="boxheading"> |
| | Related Links:</div> | | Related Links:</div> |
| | <div id="boxcontent"> | | <div id="boxcontent"> |
| - | <DL><div class="heading">Other Protocols:</div><DL> | + | <DL><div class="heading">Vitamin Notebook:</div><DL> |
| - | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Notebook/DNAAssayProtocol">DNA Protocols Page 2</a> | + | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Notebook/BCAssay">Beta-Carotene Assay</a> |
| - | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Notebook/VitProtocol">Vitamin Assays</a> | + | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Notebook/DoughMed">Dough Media</a> |
| - | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Notebook/BBProtocol">BioBrick</a> | + | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Notebook/Baking">Baking</a> |
| - | </DL> | + | </DL></div></div> |
| | <div id="primarycontent"> | | <div id="primarycontent"> |
| | </html> | | </html> |
| | __NOTOC__ | | __NOTOC__ |
| - | ======Gene Synthesis====== | + | ======Vitamin C Biosynthesis: Gene Synthesis====== |
| | | | |
| | =====8/12/11===== | | =====8/12/11===== |
| | | | |
| | + | [[File:Prs416-cut-uncut.jpg|right|200px]] |
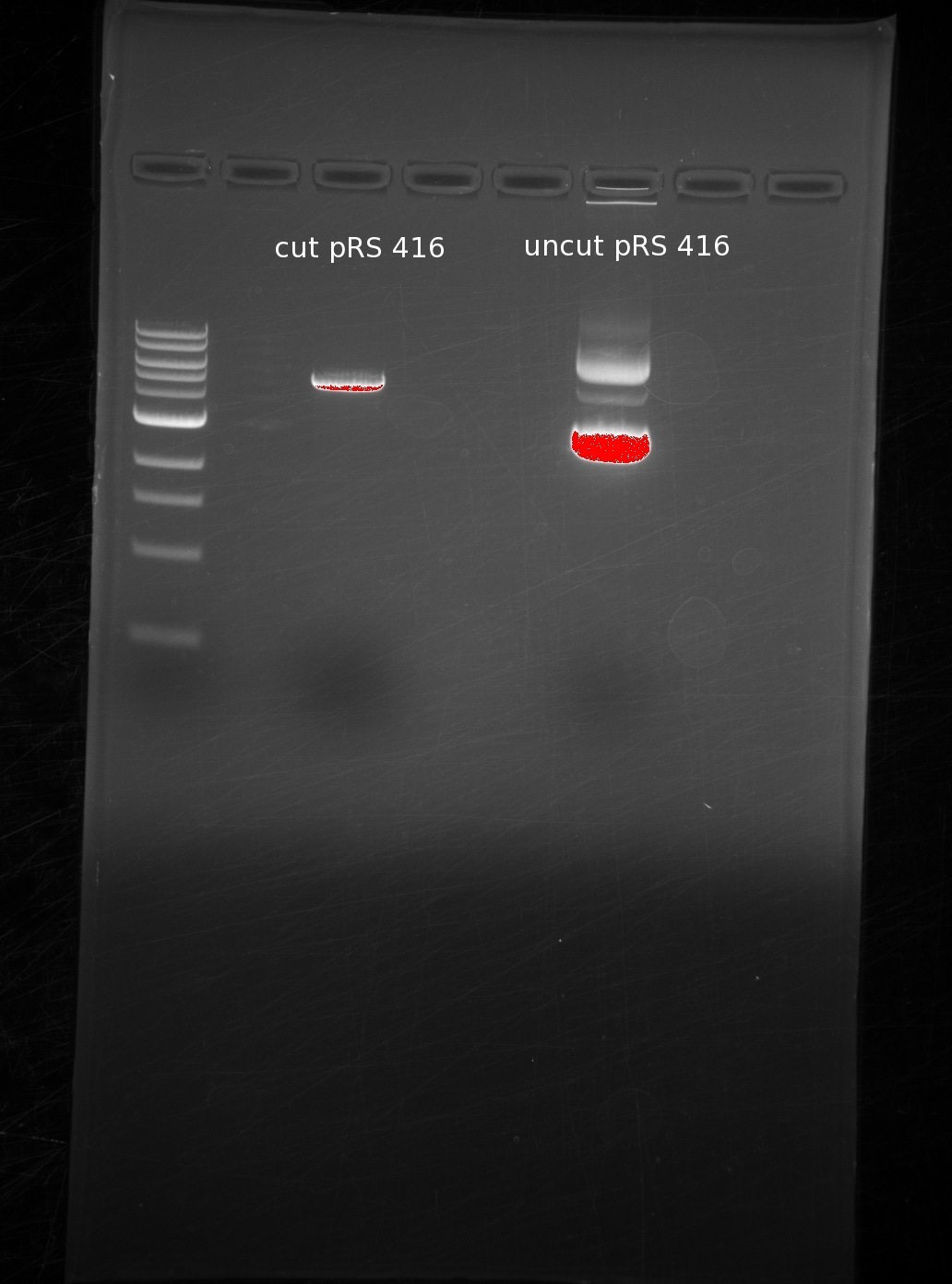
| | Our order from IDT got mixed up, so we were initially missing either the first or last oligos of GDP-mannose 3,5-epimerase and GDP-L-galactose phosphatase, respectively. Instead of working on them initially, we decided to digest our vector, which was necessary based on the desired overlaps with our genes. | | Our order from IDT got mixed up, so we were initially missing either the first or last oligos of GDP-mannose 3,5-epimerase and GDP-L-galactose phosphatase, respectively. Instead of working on them initially, we decided to digest our vector, which was necessary based on the desired overlaps with our genes. |
| | | | |
| | We digested around the BamHI and HindIII sites of pRS416, and used sequential digests because the digestion buffers are incompatible. First BamHI and then HindIII. We ran the results of our cut (1st lane) and uncut (2nd lane) in a gel for comparison. | | We digested around the BamHI and HindIII sites of pRS416, and used sequential digests because the digestion buffers are incompatible. First BamHI and then HindIII. We ran the results of our cut (1st lane) and uncut (2nd lane) in a gel for comparison. |
| - |
| |
| - | {{:lab:prs416-cut-uncut.jpg|}}
| |
| | | | |
| | The single band in the first lane represents the digestion of the vector into a linearized piece of DNA. The laddering in the uncut piece is common due to differing amounts of coiling in the circular DNA. | | The single band in the first lane represents the digestion of the vector into a linearized piece of DNA. The laddering in the uncut piece is common due to differing amounts of coiling in the circular DNA. |
| | | | |
| - | =====8/14/11=====
| |
| | | | |
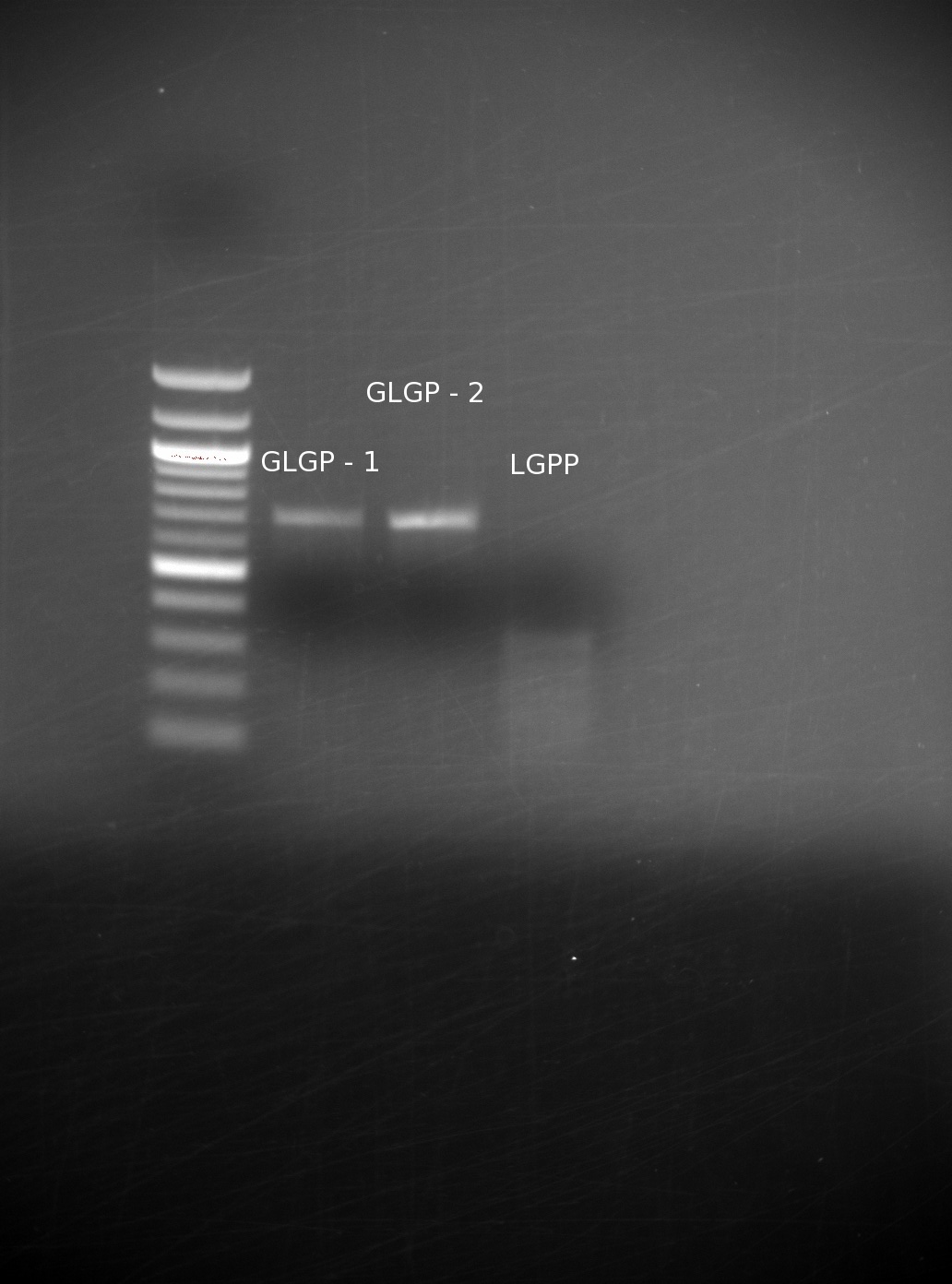
| - | We had received the missing oligos from IDT. After having gone through several failed attempts, with gels being run with no bands of the correct size, we finally achieved a set of good results. We believe that the incorrect bands were a result of not adding in the all of the appropriate oligos: even missing one can drastically We assembled them using our templateless PCR method, then ran the results of the GLGP (1st and 2nd) building blocks, and the LGPP gene on a gel
| |
| | | | |
| - | {{:lab:vitc_glgp_lgpp_tpcr.jpg|}}}
| + | =====8/14/11===== |
| | + | |
| | + | [[File:Vitc glgp lgpp tpcr.jpg|right|200px]] |
| | + | We had received the missing oligos from IDT. After having gone through several failed attempts, with gels being run with no bands of the correct size, we finally achieved a set of good results. We believe that the incorrect bands were a result of not adding in the all of the appropriate oligos: even missing one can drastically We assembled them using our templateless PCR method, then ran the results of the GLGP (1st and 2nd) building blocks, and the LGPP gene on a gel. |
| | | | |
| | Our GLGP building blocks are of the right size, however our LGPP still seems unsuccessful. We expected to have trouble with this, because we are assembling the entire part at once (as opposed to in separate building blocks), and it is around 840 base pairs including the added overlaps. | | Our GLGP building blocks are of the right size, however our LGPP still seems unsuccessful. We expected to have trouble with this, because we are assembling the entire part at once (as opposed to in separate building blocks), and it is around 840 base pairs including the added overlaps. |
| | + | |
| | + | |
| | | | |
| | =====8/17/11===== | | =====8/17/11===== |
| | | | |
| | + | [[File:Vitc lgpp gme bb.jpg|right|200px]] |
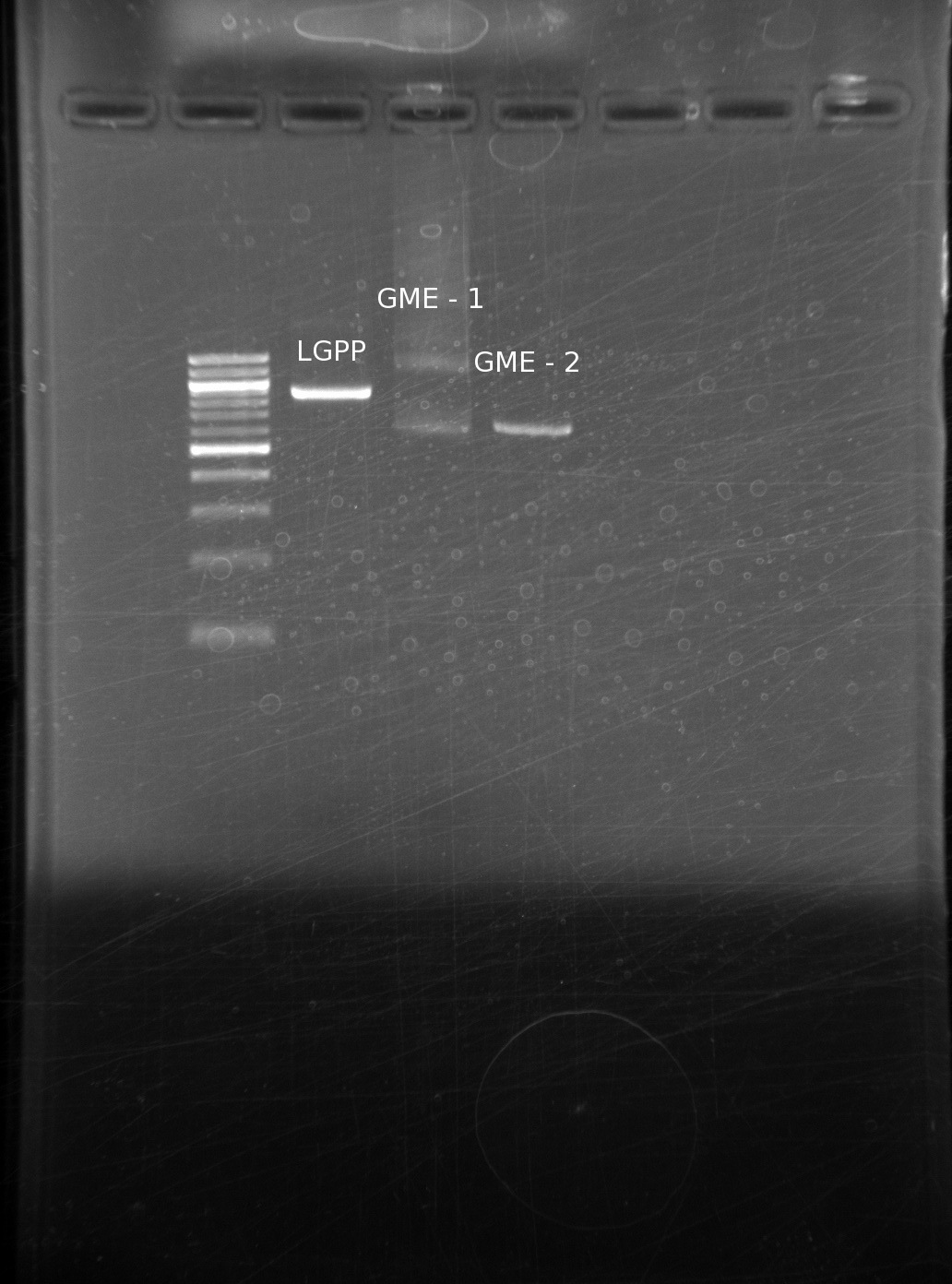
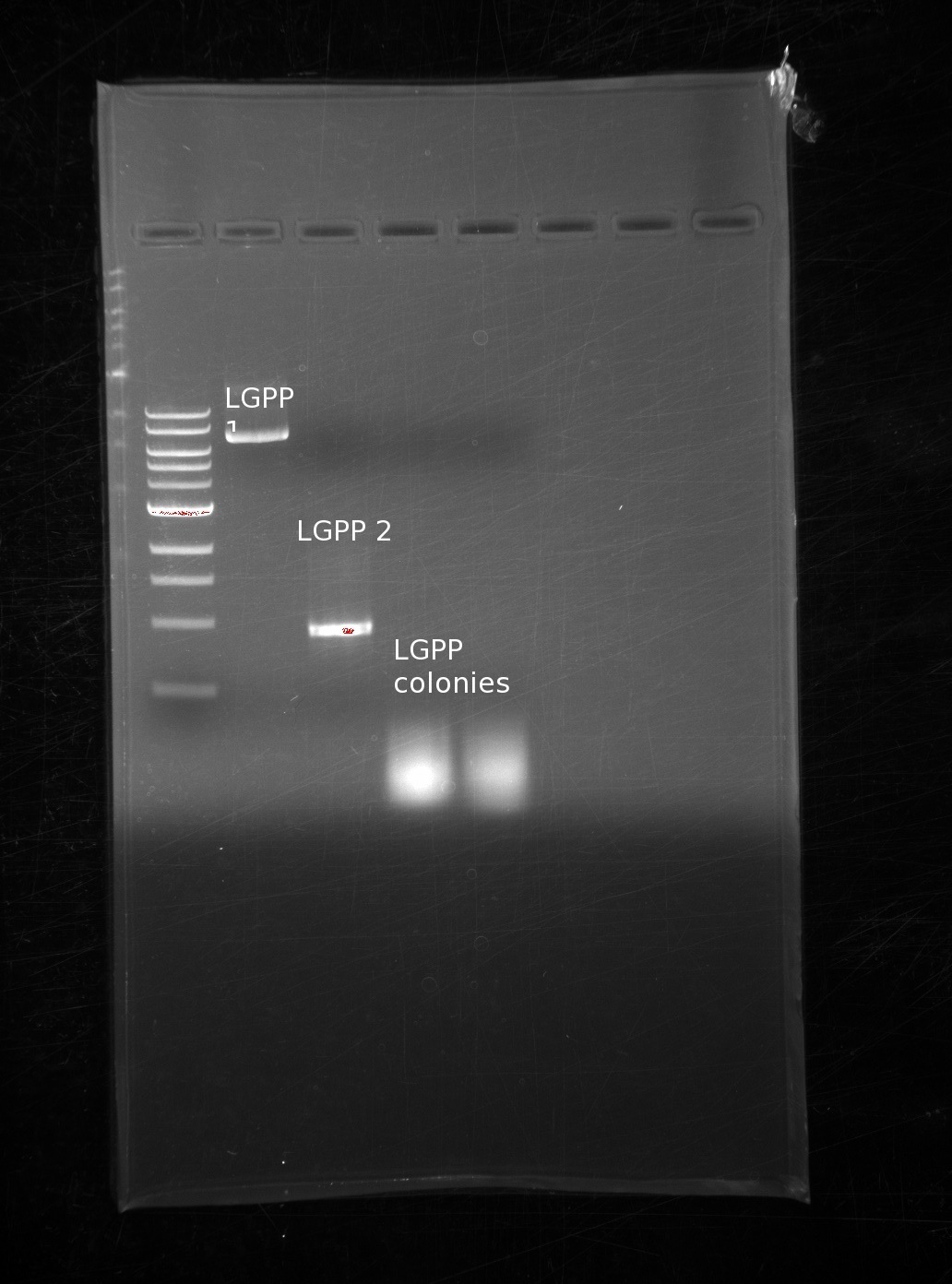
| | We have GLGP, so we will strive to make our GME and LGPP building blocks. Using the same tPCR method, we run the results on another gel (ladder, LGPP, GME 1 and 2) | | We have GLGP, so we will strive to make our GME and LGPP building blocks. Using the same tPCR method, we run the results on another gel (ladder, LGPP, GME 1 and 2) |
| - |
| |
| - | {{:lab:vitc_lgpp_gme_bb.jpg|}}
| |
| | | | |
| | The bands on every one of the genes is the appropriate size. There is some laddering on the GME 1 building block, however we do not think this is a problem. During a CPEC reaction, the only band that will be able to put the plasmid together is | | The bands on every one of the genes is the appropriate size. There is some laddering on the GME 1 building block, however we do not think this is a problem. During a CPEC reaction, the only band that will be able to put the plasmid together is |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | | | |
| | | | |
| Line 52: |
Line 77: |
| | =====8/23/11===== | | =====8/23/11===== |
| | | | |
| | + | [[File:Vitc cpcr of glgp gme.jpg|right|300px]] |
| | Unfortunately, we received no correct sequences. We realize now that it is not the PCR reactions that introduce the most errors, but rather the errors present in the oligo mixtures to begin with. On top of that, the LGPP reactions did not work at all. | | Unfortunately, we received no correct sequences. We realize now that it is not the PCR reactions that introduce the most errors, but rather the errors present in the oligo mixtures to begin with. On top of that, the LGPP reactions did not work at all. |
| | | | |
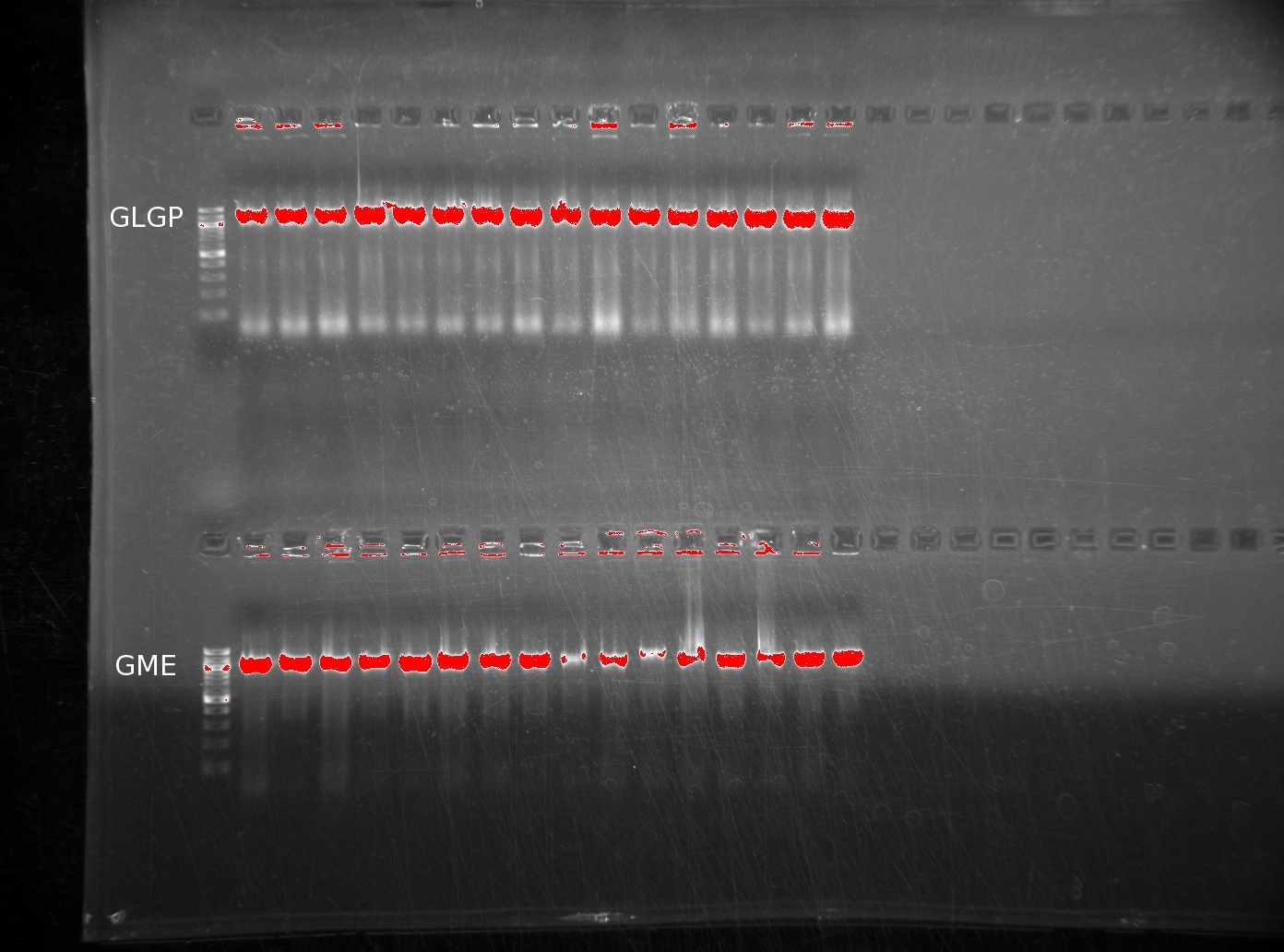
| | The lack of reactions working led us to try running a colony PCR. For colony PCR, we took 16 colonies from each plate and then performed a PCR reaction with the first and last oligos. The products were ran on 0.8% agarose gels. The first image shows GLGP (top) and GME (bottom). | | The lack of reactions working led us to try running a colony PCR. For colony PCR, we took 16 colonies from each plate and then performed a PCR reaction with the first and last oligos. The products were ran on 0.8% agarose gels. The first image shows GLGP (top) and GME (bottom). |
| - |
| |
| - | {{:lab:vitc_cpcr_of_glgp_gme.jpg|}}
| |
| | | | |
| | It is clear that these inserts are present in the colonies we've plated. However, the LGPP gel is more discouraging, returning no positive results. | | It is clear that these inserts are present in the colonies we've plated. However, the LGPP gel is more discouraging, returning no positive results. |
| Line 62: |
Line 86: |
| | =====8/26/11===== | | =====8/26/11===== |
| | | | |
| | + | [[File:Vitc lg1pp c-oligo test.jpg|right|200px]] |
| | Because of the lack of colonies on our LGPP, we looked at our fPCR products for the building blocks. We had made several trials of LGPP building blocks, and narrowed it down to two potential candidates. In addition, we decided to take 2 more colonies from the and used the same reaction. We also included 2 colonies in this next gel in the last two columns: | | Because of the lack of colonies on our LGPP, we looked at our fPCR products for the building blocks. We had made several trials of LGPP building blocks, and narrowed it down to two potential candidates. In addition, we decided to take 2 more colonies from the and used the same reaction. We also included 2 colonies in this next gel in the last two columns: |
| - |
| |
| - | {{:lab:vitc_lg1pp_c-oligo_test.jpg|}}
| |
| | | | |
| | One of our LGPP fPCR products is of the correct size, the other is far too small, and different colonies still yield nothing. It is entirely possible that we used an incorrect PCR product, however we do not know why the plasmids were taken up even though they should have remained linear. | | One of our LGPP fPCR products is of the correct size, the other is far too small, and different colonies still yield nothing. It is entirely possible that we used an incorrect PCR product, however we do not know why the plasmids were taken up even though they should have remained linear. |
| | + | |
| | + | |
| | + | |
| | | | |
| | =====8/29/11===== | | =====8/29/11===== |
| Line 104: |
Line 130: |
| | The three genes will be in the following vectors flanked by the following promoters and 3'UTRs (found in yeast) | | The three genes will be in the following vectors flanked by the following promoters and 3'UTRs (found in yeast) |
| | | | |
| - | - GLGP - pRS413, flanked by TDH3 promoter and 3'UTR
| + | # GLGP - pRS413, flanked by TDH3 promoter and 3'UTR |
| - | - GME - pRS415, flanked by RPS8B promoter and 3'UTR
| + | # GME - pRS415, flanked by RPS8B promoter and 3'UTR |
| - | - LGPP - prs416, flanked by RPL24A promoter and 3'UTR
| + | # LGPP - prs416, flanked by RPL24A promoter and 3'UTR |
| | | | |
| | We wish to use a CPEC reaction to put all of our pieces together, but first we have to add appropriate common overhangs to corresponding pieces. To this end, we have designed primers that should be able to amplify our pieces from plasmids received from our teammates with the appropriate promoter or 3'UTR region, as well as the vectors and genes. | | We wish to use a CPEC reaction to put all of our pieces together, but first we have to add appropriate common overhangs to corresponding pieces. To this end, we have designed primers that should be able to amplify our pieces from plasmids received from our teammates with the appropriate promoter or 3'UTR region, as well as the vectors and genes. |
| Line 115: |
Line 141: |
| | | | |
| | =====9/15/11===== | | =====9/15/11===== |
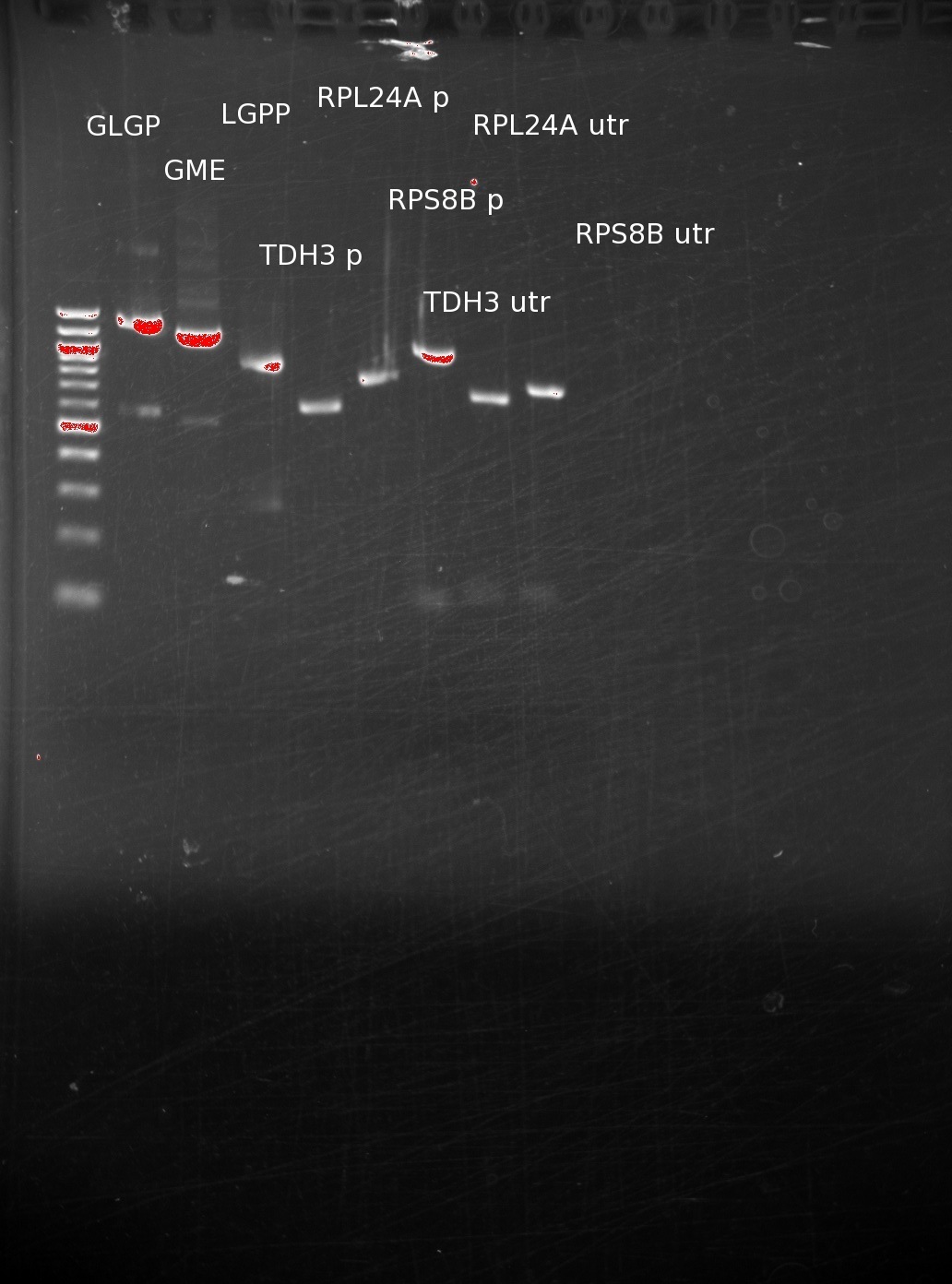
| - | | + | [[File:Vitc cpec2 overhangs.jpg|right|200px]] |
| | The primers have arrived, and we used them on genomic DNA to extract out promoters and UTR's, in addition performing PCR on our genes. The results have been run on a gel. | | The primers have arrived, and we used them on genomic DNA to extract out promoters and UTR's, in addition performing PCR on our genes. The results have been run on a gel. |
| - |
| |
| - | {{:lab:vitc_cpec2_overhangs.jpg|}}
| |
| | | | |
| | After initially having trouble getting the reactions to work correctly, we have successfully created products of the correct size. We still are having trouble with the RPS8B 3'utr, and have since designed and ordered new primers to extract larger pieces from the genomic DNA (since we assume that junk DNA after the 3'UTR should not affect gene expression. | | After initially having trouble getting the reactions to work correctly, we have successfully created products of the correct size. We still are having trouble with the RPS8B 3'utr, and have since designed and ordered new primers to extract larger pieces from the genomic DNA (since we assume that junk DNA after the 3'UTR should not affect gene expression. |
| | | | |
| - | =====9/20/11====== | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | =====9/20/11===== |
| | | | |
| | We have received the second set of primers for the RPS8B 3'UTR, however it is still giving us trouble. After attempting yet another PCR reaction, we believe that the annealing temperature for the PCR pieces is too low for good binding. We will have to order yet another set of primers. | | We have received the second set of primers for the RPS8B 3'UTR, however it is still giving us trouble. After attempting yet another PCR reaction, we believe that the annealing temperature for the PCR pieces is too low for good binding. We will have to order yet another set of primers. |
| Line 129: |
Line 158: |
| | | | |
| | The second set of primers also didn't work. We are going to attempt a gradient PCR in order to see whether our reaction works at any annealing temperature at all. We have, however, put both GLGP and LGPP into their respective CPEC vectors and plan on growing them up in ''E. coli'' before transferring them to yeast. | | The second set of primers also didn't work. We are going to attempt a gradient PCR in order to see whether our reaction works at any annealing temperature at all. We have, however, put both GLGP and LGPP into their respective CPEC vectors and plan on growing them up in ''E. coli'' before transferring them to yeast. |
| | + | |
| | + | |
| | + | |
| | + | ======Tagging Genes, Transformation, Fluorescence Microscopy, and Protein Purification====== |
| | + | |
| | + | =====9/03-04/11===== |
| | + | |
| | + | Performed 1st round PCR to create a PCR product that contains the tag sequence. There was some DNA streaking as a result of a non-optimal annealing temperature creating partial product. Performed PCR clean up and ran gel electrophoresis to confirm the success of the PCR. |
| | + | |
| | + | |
| | + | =====9/10/11===== |
| | + | |
| | + | Performed 2nd round PCR to create a PCR product that contains the tag sequence with enzymatic genes. There was minimal DNA streaking as the annealing temperature was dropped from 55C to 50C. Performed PCR clean up and ran gel electrophoresis to confirm the success of the PCR. |
| | + | |
| | + | =====9/16-17/11===== |
| | + | |
| | + | Performed Transformation using Transformation Protocol 2 and replica plated the next day. Colonies grew in a thick lawn after the first overnight incubation due to a high OD to increase the amount of competent cells. Colonies grew on each of the different strain plates. |
| | + | |
| | + | =====9/21/11===== |
| | + | |
| | + | Fluorescence Microscopy was performed on the 4 strains of transformed yeast cells. All controls showed no red fluorescence, whereas all strains showed red fluorescence. Green fluorescence was noted as beta carotene in cells. DIC microscopy showed beta carotene in lipid droplets. |
| | + | |
| | + | =====9/22/11===== |
| | + | |
| | + | Genomic Prep of the DNA tagged genes from cells was performed for protein purification assay. |
| | | | |
| | | | |
Vitamin C Biosynthesis: Gene Synthesis
8/12/11
Our order from IDT got mixed up, so we were initially missing either the first or last oligos of GDP-mannose 3,5-epimerase and GDP-L-galactose phosphatase, respectively. Instead of working on them initially, we decided to digest our vector, which was necessary based on the desired overlaps with our genes.
We digested around the BamHI and HindIII sites of pRS416, and used sequential digests because the digestion buffers are incompatible. First BamHI and then HindIII. We ran the results of our cut (1st lane) and uncut (2nd lane) in a gel for comparison.
The single band in the first lane represents the digestion of the vector into a linearized piece of DNA. The laddering in the uncut piece is common due to differing amounts of coiling in the circular DNA.
8/14/11
We had received the missing oligos from IDT. After having gone through several failed attempts, with gels being run with no bands of the correct size, we finally achieved a set of good results. We believe that the incorrect bands were a result of not adding in the all of the appropriate oligos: even missing one can drastically We assembled them using our templateless PCR method, then ran the results of the GLGP (1st and 2nd) building blocks, and the LGPP gene on a gel.
Our GLGP building blocks are of the right size, however our LGPP still seems unsuccessful. We expected to have trouble with this, because we are assembling the entire part at once (as opposed to in separate building blocks), and it is around 840 base pairs including the added overlaps.
8/17/11
We have GLGP, so we will strive to make our GME and LGPP building blocks. Using the same tPCR method, we run the results on another gel (ladder, LGPP, GME 1 and 2)
The bands on every one of the genes is the appropriate size. There is some laddering on the GME 1 building block, however we do not think this is a problem. During a CPEC reaction, the only band that will be able to put the plasmid together is
Sequencing
8/21/11
The past week had been spent researching and designing the proper reactions for putting everything together with CPEC. We have experimented since then in the lab with optimizing the protocol, until we have created one that successfully creates the desired plasmid.
We used CPEC to insert each gene into a vector (pRS 416), and then transformed these vectors via electroporation into E. coli. pRS 416 has an expressed gene for ampicillin resistance, so we can grow up the E.coli on ampicillin plates. We grew up the cells overnight, selected a single colony, and then grew that colony up in liquid media. We obtained plasmid DNA from these cultures using the QIAgen miniprep protocol, and sent these in to be sequenced.
8/23/11
Unfortunately, we received no correct sequences. We realize now that it is not the PCR reactions that introduce the most errors, but rather the errors present in the oligo mixtures to begin with. On top of that, the LGPP reactions did not work at all.
The lack of reactions working led us to try running a colony PCR. For colony PCR, we took 16 colonies from each plate and then performed a PCR reaction with the first and last oligos. The products were ran on 0.8% agarose gels. The first image shows GLGP (top) and GME (bottom).
It is clear that these inserts are present in the colonies we've plated. However, the LGPP gel is more discouraging, returning no positive results.
8/26/11
Because of the lack of colonies on our LGPP, we looked at our fPCR products for the building blocks. We had made several trials of LGPP building blocks, and narrowed it down to two potential candidates. In addition, we decided to take 2 more colonies from the and used the same reaction. We also included 2 colonies in this next gel in the last two columns:
One of our LGPP fPCR products is of the correct size, the other is far too small, and different colonies still yield nothing. It is entirely possible that we used an incorrect PCR product, however we do not know why the plasmids were taken up even though they should have remained linear.
8/29/11
We have decided to make new CPEC reactions for each genes as well as LGPP, and then send 16 different colonies as opposed to only one. These colonies were grown up overnight in liquid media (with ampicillin resistance to ensure only E. coli with our (and replica plated, so we would have more colonies should they become necessary), mini-prepped, and then an aliquot of the mini-prepped DNA was sent with custom sequencing primers.
9/1/11
We obtained no positive results. In addition, we also had a high rate of the sequencing reactions failing. We believe the cause was degradation/other problems with our custom sequencing primers.
We examined the vector's sequence, and found that the T7 promoter primer and the M13Reverse primer (both universal sequencing primers) should give us the forward and reverse sequences of our genes in the vector. We also realized that instead of spending a large amount of time growing and mini-prepping many different colonies, we could instead plate the same colony on two separate plates, keeping one and sending the other in to be sequenced.
The plates would be labeled in a grid, and have each colony streaked in the appropriate section of the grid. This way, the gene-sequencing service would be able to perform the sequencing reaction with a bacterial sample rather than a DNA sample, and once the sequencing reactions got back, we would be able to selectively mini-prep the winners we streaked out on replica plates.
Since GLGP is the longest reaction gene, we decided to use our middle sequencing primer to ensure that we got the whole gene, and because our second custom primer should cover the rest of the gene, we will not request an additional M13Reverse primer reaction
9/5/11
Upon getting the results back, we found that only LGPP had given us a positive result. We would need to send in more colonies of GME. For GLGP, only one sequence returned no errors using the T7 primer (the first 700 base pairs) unfortunately, ALL of our custom primer reactions didn't work. However, now that we have a certain result for a perfect LGPP gene, we will focus our attention on GLGP and GME.
We took 16 more single colonies of each and streaked them out on two separate plates.
9/7/11
Today we received our sequencing reactions back, and have determined that we have all of our genes properly formed.
Synthesizing Vitamin C Yeast
9/10/11
In order to create a strain of yeast that has all three of the genes in it, we cannot use the same vector for all 3 genes, as we have been in the past. The reason for this is that we cannot use the same selectable marker to choose the yeast cells which have all 3 plasmids, since they would all have the same marker. In other words, we could not tell which yeast colonies grew up with one, two, or all three plasmids.
In order to counteract this problem, we have decided to put the genes into 3 separate vectors: we will use pRS413 and pRS415 in addition to pRS 416. These vectors correspond to his-, leu-, and ura- respectively. This way we can make a plate lacking these three metabolites, and any cells that grow on them will have to have all three plasmids.
But this will only get the genes inside of the yeast, it will not necessarily express them. Therefore, we need to make a proper cassette, with a promoter region in front of the gene and a 3'UTR trailing it.
The three genes will be in the following vectors flanked by the following promoters and 3'UTRs (found in yeast)
- GLGP - pRS413, flanked by TDH3 promoter and 3'UTR
- GME - pRS415, flanked by RPS8B promoter and 3'UTR
- LGPP - prs416, flanked by RPL24A promoter and 3'UTR
We wish to use a CPEC reaction to put all of our pieces together, but first we have to add appropriate common overhangs to corresponding pieces. To this end, we have designed primers that should be able to amplify our pieces from plasmids received from our teammates with the appropriate promoter or 3'UTR region, as well as the vectors and genes.
9/11/11
The vectors have been successfully linearized, along with addition of the appropriate overlaps. The primers, however, didn't work on the promoter and 3'UTRs. Because of that, we have designed new ones that will instead both take the promoter and UTR regions directly out of genomic DNA and add the necessary overlaps in one reaction.
9/15/11
The primers have arrived, and we used them on genomic DNA to extract out promoters and UTR's, in addition performing PCR on our genes. The results have been run on a gel.
After initially having trouble getting the reactions to work correctly, we have successfully created products of the correct size. We still are having trouble with the RPS8B 3'utr, and have since designed and ordered new primers to extract larger pieces from the genomic DNA (since we assume that junk DNA after the 3'UTR should not affect gene expression.
9/20/11
We have received the second set of primers for the RPS8B 3'UTR, however it is still giving us trouble. After attempting yet another PCR reaction, we believe that the annealing temperature for the PCR pieces is too low for good binding. We will have to order yet another set of primers.
9/23/11
The second set of primers also didn't work. We are going to attempt a gradient PCR in order to see whether our reaction works at any annealing temperature at all. We have, however, put both GLGP and LGPP into their respective CPEC vectors and plan on growing them up in E. coli before transferring them to yeast.
Tagging Genes, Transformation, Fluorescence Microscopy, and Protein Purification
9/03-04/11
Performed 1st round PCR to create a PCR product that contains the tag sequence. There was some DNA streaking as a result of a non-optimal annealing temperature creating partial product. Performed PCR clean up and ran gel electrophoresis to confirm the success of the PCR.
9/10/11
Performed 2nd round PCR to create a PCR product that contains the tag sequence with enzymatic genes. There was minimal DNA streaking as the annealing temperature was dropped from 55C to 50C. Performed PCR clean up and ran gel electrophoresis to confirm the success of the PCR.
9/16-17/11
Performed Transformation using Transformation Protocol 2 and replica plated the next day. Colonies grew in a thick lawn after the first overnight incubation due to a high OD to increase the amount of competent cells. Colonies grew on each of the different strain plates.
9/21/11
Fluorescence Microscopy was performed on the 4 strains of transformed yeast cells. All controls showed no red fluorescence, whereas all strains showed red fluorescence. Green fluorescence was noted as beta carotene in cells. DIC microscopy showed beta carotene in lipid droplets.
9/22/11
Genomic Prep of the DNA tagged genes from cells was performed for protein purification assay.
 "
"