Team:Imperial College London/Extras/Brainstorming
From 2011.igem.org
Brainstorming
This page contains a summary of the ideas we developed throughout our brainstorming sessions towards the beginning of the project.
Bacterial sunscreen
- Evidence suggests that several components found in most sunscreens are harmful to us and can be carcinogenic, also most sunscreens only protect against UV B (315-280 nm) and not UV A- These sunscreens use metal oxides (Zinc oxide) to absorb UV radiation, but the effects of absorbing these metals into your skin are not fully understood and are thought to lead to production of reactive oxygen species and could lead to melanomas rather than preventing them.
- Scytonemin is a pigment found in cyanobacteria which protects them from UV radiation, absorbing 325-425nm. Its synthesis requires three enzymes, SycA-C
Mycosporine-like amino acids (MAAs) are produced by organisms adapted to environments with high levels of sunlight (eg. cyanobacteria and algae), protecting them from UV radiation. There are 20 types and they also serve as anti-oxidants by stabilising free radicals (anti-ageing?). In a bioinformatics study the genes YP_324358 (predicted DHQ synthase) and YP_324357 (O-methyltransferase) were identified in A. variabilis PCC 7937 cyanobacteria. (http://www.sciencedirect.com/science/article/pii/S0888754309002353) MAAs have already been recognised to have sunscreen potential and are found in some anti-wrinkle creams
- gene cluster encoding 4 enzymes required for all MAA synthesis. Expressed this gene cluster from a cyanobacterium into E. coli and got pigment production. (Text by Nikki)
Prodigiosin pigment
- UV protecting properties- synthesis controlled by quorum sensing
- absorbs in UV range from 240 to 400 nm as well as in the visible spectrum from 400 to 600 nm
- also antibiotic and anti-cancer (induces NAG-1 pro-apoptotic gene in human breast cancer cells)
- Prodigiosin biosynthesis gene cluster (pig cluster) contains ~15 ORFs in Serratia strains (Harris et al., 2004)
- PigS and PigP regulate prodigiosin biosynthesis in Serratia (Gristwood et al., 2011)
- Streptomyces griseoviridis 2464-S5 produces prodigiosin R1, gene cluster of 24 open reading frames, including 21 genes (rphD-rphZ) homologous to prodigiosin biosynthesis genes in the red cluster in Streptomyces coelicolor A3(2). The expression of rphN in S. coelicolor lacking redN restored the production of prodigiosin (Kawasaki et al., 2009)
- could also be used as a dye in clothing. pigment production from microorganisms by large-scale fermentation would be environmentally friendly and sustainable. Could make clothes with inherent UV protection, but they would all be red.... (Text by Nikki)
Sci-Fi prodigiosin ideas (developed by Rebekka, Nick and CJ from the RCA):
-use prodigiosin (red pigment) as the new "colour of health" (know something is sterile rather than assume it is)
-possible future uses: in decontamination/ as a "panic room"/ sterile hospice
-possible future products: hand gel, clothing (e.g. protective suits in bioreactor plants), decontamination paint (in hospitals etc)
-actual uses: anti-cancer (could be in a red drip, red pill?), anti-malarial (drug verification because pigment colour is hard to fake)
Eventually, this idea was scratched because optimising the production pathway does not contain enough synthetic biology. In addition, the compound is immunosuppressive (http://pubs.rsc.org/en/Content/ArticleHtml/2008/CC/b719353j) and would therefore be disadvantageous to normal people. Many of the envisaged applications would only work with less problematic analogues of prodigiosin.
Synthetic graphite
- Synthetic Graphite normally has a high electrical conductivity than the natural ones.- the process involves turning amorphous carbon into crystal under extremely high temperature.
- no evidence shows that carbon(element) can be either iuput or output of bacterial metabolism.
- therefore, we can pass this topic (Text by Nina)
Anti-venom
- traditional way of anti-venom production:1. injecting venom or detoxified venom into a horse (tiny amount, multiple times)
2. after the antigen growing period, contract the horse blood plasma
3. use stomach digestive enzyme to breakdown the anti-venom protein into smaller globin
4. use (NH4)2SO4 to salt out the globin (purification) [1]
- oxides like K2MnO4 can neutralise venom by denaturing the polypeptide chains
- complex ligase like AuCl2 can denature venom by binding with them, when preventing venom entering the tissues [2]
-immune system has a stronger response to venom [3]. When the mast cells are stimulated, they release histamine. Histamine can subside the venom
- histamine producing bacteria: found in tuna; 18 types of bacteria such as Clostridium perfringens and other anaerobic bacteria
- to conclude:
1. decide the working mechanism (toxicology) of a specific type of venom: does it attack neurons? brain cells? cardiovascular system? respiratory system?
2. does it react with histamine?
3. bacterial production of histamine
REFERENCES:
[1]http://en.wikipedia.org/wiki/Antivenom
[2] http://life.91sqs.com/html/zazhi/yixueyushehui/2011/0113/1453.html
[3] “ Development of antibody against Naja naja atra venom using phage display and single-chain Fv antibody technology ” Master Graduation Paper NUK http://ethesys.nuk.edu.tw/ETD-db/ETD-search-c/view_etd?URN=etd-0825110-170246 (Text by Nina)
We discussed the anti-venom idea with Koby (one of the RCA students). He helped us expand the idea to maybe provide protection against viruses as well. We even developed the idea to use a seasonal hand wash containing the purified antibodies from the season's viruses in order to create a world where a handshake would be more than just a form of greeting but also a way to pass on immunity.
RCA sci-fi story ideas:
Nuclear winter leaving people without immune systems. Use external immune system to save everybody?
Synthetic life can only use certain amino acids, firewall?
Biocomputers where man and machine are converged closer together.
Floppy disk baby?
Mock news articles (Times, science article, tabloid), comic strips (simplified version of our project), documentary type video?,
Terrorist attacks (Text by Chris)
Antibody research:
-Single-domain antibodies such as the nurse shark derived IgNAR and the camelid derived VHH have been used for many purposes and have recently started to gain popularity among the scientific community.
-Contain CD3 loop that gives these Ig's an advantage when looking for cryptic viral epitopes. However, contain ten epitope copies and might still infect. Solved by increasing their mass.
-Both of these Ig's are stable enough to be administered orally.
-Orally administered transformed lactobacilli were used to administer anti-TNF Ig.
-Small dimensions of VHHs allow it to be easily tagged.
-Lactobacilli have been engineered that produces VHH's at a rate fast enough to prevent infection by p2 bacteriophage. Possibly use lactobacillus for screening and E. coli for secretion?
-VHH's and IgNAR's have been effective in detecting poliovirus and inhibiting its replication in vitro, as well as preventing the assembly and secretion of hepatitis B. Possible to use VHH's as intrabodies vs. HIV-1?
-N-glycosylation increases stability.
-Studies demonstrated that pre-immune libraries can be used for rapid generation of Ig's against a large number of harmful antigens. Troublesome low sensitivity overcome by using phage-displayed instead of purified antibodies.[1]
References:
[1]Ario de Marco, “Biotechnological applications of recombinant single-domain antibody fragments,” Microbial Cell Factories 10, no. 1 (2011): 44. (Text by Chris)
To generate cell surface anchored antibody fragments: VHH sequence is fused to the anchor sequence from proteinase P of L. casei.
To generate the secreted VHH, stop codon after E-tag. (Text by Chris)
Sorting cells - A method is needed to sort cells by their ability to bind the venom proteins
- One such method would be to have the cells secrete their anti-venom proteins and then flood the cells with venom. The best cells would survive and the weak ones would be killed. However, this may cause the production of proteins that bind the venom components that are less dangerous such as phospholipases and oxidases. It would be more important for the anti-venom to inhibit the neuro-muscular disruptive proteins
- One effective method would be to use Fluorescence Activated Cell Sorting, which is able to sort cells by their fluorescence. See: http://www.bio.davidson.edu/courses/genomics/method/FACS.html
- This would require a system that expresses a fluorescent reporter in response to the binding of venom proteins to the cell-surface proteins, but I cannot be too specific until I know how the anti-venom proteins are going to be expressed
- Yet another method would cause the death of any cells that do not bind to the venom with high enough affinity - it would be similar to the methods employed in T-cell selection in eukaryotic immune systems, but would be more tricky as it has to occur in a bacterium
- The ideal then, would be to have all the cells that have little or no binding affinity killed, and then those that do bind express GFP so that the fluorescence can be quantified and the binding can be rated
- Again, I'd need to understand the anti-venom proteins before I could suggest any particular systems
- Perhaps a second species of bacteria could be used to express the venom proteins on their surface, and come into contact with the anti-venom producing cells, causing contact-dependant stimulation, so that those that interact by the venom-antivenom complexes will be stimulated to divide whilst the remaining cells will potentially be killed See: http://www.ncbi.nlm.nih.gov/pubmed/21085179 (Text by Frank)
In vivo mutagenesis
- we need a method for random mutagenesis of peptides to create high affinity binding proteins to the multiple components of venom
- PCR based mutagenesis is namely used for site directed mutagenesis and has several biases that make it not ideal for random mutation
- in vivo homologous recombination inherent to yeast can be exploited for protein mutagenesis
(Pirakiticulr et al., 2010)
MAGE (multiplex automated genome engineering)
See: http://www.nature.com/nature/journal/v460/n7257/fig_tab/nature08187_F4.html - a method for large scale evolution of cells
- has been used to optimize the 1-deoxy-d-xylulose-5-phosphate (DXP) biosynthesis pathway in E.coli for isoprenoid lycopene overexpression with a pool of synthetic DNA to modify 24 genes in the pathway creating > 4.3 billion combinatorial genomic variants per day (Wang et al., 2009
- can do insertions, deletions and mismatches
See: http://www.nature.com/nature/journal/v460/n7257/fig_tab/nature08187_F1.html (Text by Nikki)
However, all currently known methods of in vivo mutagenesis are not able to mutate a specific gene without requiring a transformation step. We therefore propose the following mutagenesis mechanism:
We would therefore use a novel mechanism based on an rt reaction (Text by Rebekka)
Type of venom
Venom refers to varieties of toxins produced by certain types of animals. One of the most common venoms are produced by snakes where 15% of 3,000 species of snakes are found poisonous. Snake venom consists of proteins, enzymes, substances with a cytotoxins, neurotoxins and coagulants
Most snake envenomings and fatalities are found in South Asia, South East Asia and sub-Saharan Africa with the high fatality rate of 125,000 deaths per annual. Among these India is reported the most cuased by big 4 including Russell's viper, Indian cobra, saw-scaled viper, and the common krait. Indian cobra is found the most famous and make the highest fatality rate (43%) in India and Southeast asia.
Indian cobra venom contains a potent post-synaptic neurotoxin which acts on the synaptic gaps of the nerves, thereby paralyzing muscles, and in severe bites leading to acute respiratory failure or cardiac arrest. The components of venom include lysis enzymes such as hyaluronidase which increase the spread of the venom. Its toxicity is found one of the highest based on LD50 value in mice. Symptoms of cobra envenomation can begin from 15 minutes to two hours after the bite, and can be fatal in less than an hour.
Despite the advance in emergency therapy, antivenom is often only the effective treatment. In treatment antivenom is injected into patient intravenously which could neutralize the toxin. Collecting of antivenom is done by milking the venom and injected into the cattle. The subject will undergo immune response where its antibody produced can be collected. This common method of obtaining antivenom can tackle many toxins in the venom but however is considered unproductive since only small amount of antivenom is produced from the animal blood which is due to complicated serum purification process from the animal's serum. Waiting of animal recovery from venom also make its production quite slow and in several cases the animals die after injection.
Therefore bacteria should be introduced as a substituted platform utilizing synthetic biology to deal with the problems mentioned above. Apart from existing characterised antivenom, other antivenoms could be more easily discovered using the various mutagenesis of variable region of antivenom antibody. To develop this platform, well known venom could be produced to test this platform. Indian cobra snake is therefore chosen as the first target due to its generality, high toxicity and it is also one of the highest profile antivenom discovered which could potentially save many victims from this fatal snakebite.
The way to engineer the bacteria is to mutagenise the shark antibody gene to allow different variable regions of the antibody to be expressed on the surface of the bacteria in different libraries. The venom is screened onto each plate of different bacteria libraries. The venom will bind to the right antibody and trigger the signal cascade which results in the expression of the fluorescence proteins which can be detected by FACs. machine. The bacteria that produces the antibody for the venom will be subjected to DNA sequencing which could be the platform for producing bacteria expressing antibody specific to the venom in the future. (Text by Ming)
So the method for the anti-venom generation begins as follows:
1: Take the venom proteins and bind biotin to either the C or N terminus so that it is able to be fixed to a large ferro-magnetic beads that are coated in streptavidin. Streptavidin has an extremely high affinity for biotin and the beads can be picked up with a neodynium magnet - so the bacteria with the 'antibody' on their surface that is able to bind to the venom proteins will be attached to the bead.
2: The bacteria will have a plasmid that encodes a gene for a single-chain variable fragment, a fusion protein of the variable regions of the heavy and light chains of an immunoglobulin. This will be displayed upon the surface of the bacterium and will be on a plasmid that has a very mutagenic effect as described by Rebekka.
Alkaloid isorhy against Parkinson's
- It is rumoured to be a potential treatment for Parkinson's and it would make a good project if this were to be produced by bacteria- On further research it turns out that the evidence for this drug as a treatment is weak and there is no information available about the gene or genes that encode it, so the idea was dropped (Text by Frank)
Bacterial tattoo
- After being inspired by a student from the Royal College of Arts who presented us with her work on a project to create a living dress, Frank began to research the notion of a melanin tattoo, so that alpha-melanin stimulating hormone is applied to the skin and held in place until the skin darkens in the shape of the template. The alpha-MSH could be produced by bacteria.- the alpha-MSH gene is produced as one gene that also contains beta-MSH and gamma-MSH that are made available through post-transcriptional processing, so only the alpha-MSH region is required as it is the best characterised and has been expressed before
- Once the alpha-MSH is expressed, it can be collected, soaked into silk (for example) that is cut into a pattern and will allow the hormone to diffuse into the skin, producing a (probably temporary) tattoo. From: http://en.wikipedia.org/wiki/Melanocyte-stimulating_hormone
- However, it seems that alpha-MSH is a rather powerful aphrodisiac and so a different hormone will have to be chosen, in addition, it seems that the hormone is unlikely to penetrate the skin as there are many different layers as well as proteases secreted by the skin
- alpha-MSH, or its analogues are already used as tanning solutions and the analogues are considerably stronger
- so then, bacteria could express the gene for melanotan - which is a cyclic lactyam analogue, and this will be very difficult to express, as a method will have to be found for cyclisation.
- One further problem: "As of 2010 no compound incorporating the melanotan II peptide has ever been approved for use by any governmental drug regulatory bodies outside of clinical trials. Unlicensed and untested powders sold as "melanotan II" are found on the Internet and are reported to be used by thousands of members of the general public. Multiple regulatory bodies have warned consumers the peptides may be unsafe and ineffective in usage with one regulatory agency warning that consumers who purchase any product labeled "melanotan" risk buying a counterfeit drug. Medical researchers and Clinuvel Pharmaceuticals, the company developing the related peptide afamelanotide, has warned consumers that counterfeit products sold using the names "melanotan I and II", "pose a hazard to public health"." From http://en.wikipedia.org/wiki/Melanotan
- alpha-MSH will have to be used, but this time with the novel method for crossing the skin: Transdermal iontophoresis. This is a non-invasive way for hydrophilic proteins to be transported across the skin but I do not know what kind of resolution is possible with this device. Whatever the pattern achieved, be it a nice dot or a blotted smudge, the students from the RCA will surely help to make it look pretty. (Text by Frank)
Vampiric bacteria
-The aspect of a Vampiric bacteria that is designed to get rid of blood clots produced by trauma induced clotting or during complex medical procedures is intriguing.-Expression of Hirudin is possible in systems such as E. coli. In 2007 Berkeley produced a chassis for a E. coli that could be introduced into the blood stream after inactivation.
-However, it is difficult to have the non-viable cell lysis occur in the correct location and therefore an anticoagulant could just as well be injected into the patient.
-For this to work, we would require an expression system that is able to express Hirudin (produced usually by leech salivary glands and has been successfully expressed in E. coli [1]), express anti-angiotensin (it is possible to express Fab fragments in E. coli [2]) and targeting the fibrin (can be done by expressing Tissue plasminogen activator).
-The idea would be to have the chassis recognize a blood clot or an area of damage and prevent clotting and/or clear clots. A method for having the system recognize when to secrete hirudin would be by having the bacteria sense trauma related chemokines and have the chassis secrete the protein only when it senses above a certain threshold of these chemokines or we could try to express protease-activated receptors (GPCR) that are cleaved by activated thrombin (the target of hirudin). Direct application would only benefit over the use of leeches in that the chassis is more aseptic then a leech bite. The biggest issue remains the fact that for this to work we would have to inject the patient with living E. coli that can evade the human immune system.
A new method of boosting biosynthesis has been obtained through the use of RNA scaffolds: http://www.sciencemag.org/content/early/2011/06/22/science.1206938
Reference:
[1]Shuhua Tan et al., “Efficient expression and secretion of recombinant hirudin III in E. coli using the L-asparaginase II signal sequence,” Protein Expression and Purification 25, no. 3 (August 2002): 430-436.
[2]Saad A Masri et al., “Cloning and expression in E. coli of a functional Fab fragment obtained from single human lymphocyte against anthrax toxin,” Molecular Immunology 44, no. 8 (March 2007): 2101-2106.
[3]Ji Qiu, James R. Swartz, and George Georgiou, “Expression of Active Human Tissue-Type Plasminogen Activator in Escherichia coli,” Applied and Environmental Microbiology 64, no. 12 (December 1998): 4891-4896. (Text by Chris)
Converting fallen leaves into useful products
-Aquatic hyphomycetes has been recognized as critical for controlling the process of leaf litter breakdown. The activity of this fungus is affected by C:N ratio, lignin content, pH of water, temperature, and abundance of nutrients (i.e. O2). They produce B-glucosidase, Cellobiohyhrolase (cbhI family), B-xylosidase (xlnR) and phenoloxidase (Pox2) to promote leaf degradation.-As leaves decay, they produce heat. Leaves will decompose into an excellent organic soil amendment that can be used as a soil conditioner.
-The decomposition process is slow (i.e. leaves require 5 months to 2 years to decompose), could combine with Nick’s gene expression amplification?
-However, rapid decomposition would consume a large amount of O2 and create anaerobic conditions. Could we engineer all these into facultative anaerobes?
References: [1] FEMSMicrobiolLett 264(2006)246–254, DOI:10.1111/j.1574-6968.2006.00462.x
[2] E.N. Tamayo et al. / Fungal Genetics and Biology 45 (2008) 984–993, doi:10.1016/j.fgb.2008.03.002
[3] APPLIED AND ENVIRONMENTAL MICROBIOLOGY, June 2008, p. 3481–3489, doi:10.1128/AEM.02893-07
[4] Mutagenesis Advance Access published June 15, 2006, doi:10.1093/mutage/gel025
[5] http://herbarium.usu.edu/fungi/funfacts/decay.htm
[6] http://onlinelibrary.wiley.com/doi/10.1002/iroh.201111355/pdf (Text by Yuanwei)
Fuel from food waste
-Microbes in food waste like heterotrophs, cyanobacteria, microalgae and purple bacteria produce biohydrogen. Hydrogen has more potential energy than petrol. Hence, food waste can be turned into valuable energy. Fermentative bacteria use carbohydrates like sugar to produce hydrogen and acids. Purple bacteria, use light to produce energy (photosynthesis) and make hydrogen to help them break down molecules such as acids. http://www.sciencedaily.com/releases/2008/07/080716204805.htm-Hydrogen is produced by feeding waste products from a chocolate factory to Escherichia coli bacteria. E coli ferments the sugars in the chocolate waste, which generated organic acids so toxic to the bacteria that they began converting formic acid to hydrogen. http://environment.about.com/od/renewableenergy/a/chocolatefuel.htm
-Cellulose waste can be converted to energy by using enzyme cellulase. The gene that codes for cellulase has been isolated and grown in large quantities by E. coli. A number of photosynthetic bacteria, nonphotosynthetic bacteria, cyanobacteria, and green, red, and brown algae produced the enzyme hydrogenase, which is necessary to make hydrogen. http://www.accessexcellence.org/RC/AB/BA/Future_Fuel.php (Text by Yuanwei)
Feather-Eating Bacteria
Bacillus licheniformis strain PWD-1 breaks down feathers into a feather-lysate compound. Feather-lysate provides a low-cost, highly digestible protein source for livestock feed. Bacillus has also been shown to secrete a keratinase enzyme that hydrolyzes proteins such as collagen, elastin, and keratin. Potential application in breakdown of livestock carcasses. The gene encoding the enzyme keratinase of Bacillus licheniformis is '''kerA'''. http://www.accessexcellence.org/RC/AB/BA/The_Smell_of_Wealth.php, "http://aem.asm.org/cgi/content/abstract/61/4/1469 (Text by Yuanwei)Turning C3 plants into C4 plants
problem: How to make C3 plant operating in sunny and arid areas or how to reduce photorespirationsolution: Create a bacteria which penetrates plant cells, creates high concentration of HCO3− and packages it into vesicles, inactive Carbonic anhydrase is added to the vesicles, releases vesicles with chloroplast localisation signal, releases vesicles into the chloroplast, upon fusion CA is activated and changes HCO3− into carbon dioxide, which is then highly concentrated in a chloroplast and reduces rate of O2 binding to the Rubisco simply by increasing concentration of CO2.
Chassis: E.coli or Sinorhizobium meliloti
Bacterial infection: Nod factors
Bacteria of Rhizobium spp. are capable of infecting a plant and forcing it to develop an extra organ - Nodule, where these bacteria then intracellularly (a bit like organelles) reside. They do this to develop a mutualistic relationship with plant. We could use this mechanism of infection and acceptance by using the entire "Nod box" a cluster of genes involved in signalling to the plant to allow entry through the specially deformed root (induced by the Nod factors) or by crack entry. Each of the two mechanisms involves plant release of the flavonoids in the first place to trigger the Nod factors in the first place.
File:Rhizobium.gif
problems:
-A lot of plants do not have Nod factor receptors, as wild type Rhizobium infects only legumes, so we would have been restricted to legumes as well. Also there is specificity among different Nod factors and their receptors on the plants meaning that not every Nod box containing bacteria could infect every plant.
-In theory inserting a whole "Nod box" of genes into E. coli should enable E.coli to function in relation to the plant much in the same way as Rhizobium does, however we can not be sure of that, even though there is evidence that some genes in Rhizobium (NodD) have orthologues in E. coli (glmS).
-Plant accepts Rhizobium as a symbiont and expects to get something from it, therefore if we were to use Rhizobium as chassis we could leave the initial nitrogenase function intact, however there might be a problem using E. coli as it would not be capable of fixing nitrogen the plant might not accept its infection thread.
-Rhizobium forces plant to form nodule on the root, however ideally we would want to set up infection into the leaves. Maybe possibility to send vesicles through the xylem to the leaves, however vesicles would face problem of crossing plant cell wall.
Accumulation of HCO3− and packaging into the vesicles: CaA and carboxysome
A lot of cyanobacteria / algae, use specialised carboxysomes to accumulate HCO3− through a number of HCO3− transporters and carbon dioxide converting enzyme Carbonic anhydrase which performs interconversion of CO2 and HCO3−. Different genes in C. reinhardtii (cupA, cupB) act as transporters of CO2 and automatically convert it to HCO3−. There is a number of other transporters utilised by cyanobacteria, but these just transport HCO3−
and do not convert it to CO2, and therefore are not useful to us. Then a number of genes involved in carboxysome production would have to be included in the chassis as well. Also normal carboxysome in a cyanobacterium contains a number of other protein products to convert CO2, however these are not necessary as carbon fixation would be performed by the plant itself. Finally a CaA - carbonic anhydrase converting HCO3− to CO2 would be included, also Cso3 a Carbonic anhydrase embedded in the carboxysome membrane would be present.
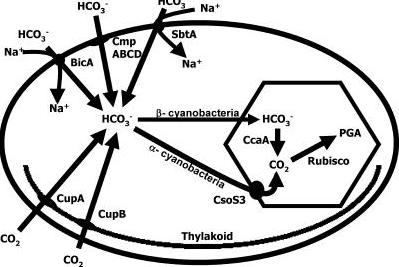
However it needs to be inactive within the carboxysome/vesicle and active only upon entry into chloroplast. Therefore possible fusion protein with 3 domains could be created containing CA on the inner end, then transmembrane subunit and a transit/fusion peptide targeting it to the chloroplast. Upon fusion into chloroplast the fusion protein would be cleaved and CA would become active.
problems:
-Creation of carboxysome ( a whole "organelle") within a chassis not previously having any.
-Creating vesicles out of carboxysome, which would not release any of its content out into bacterial cytoplasm (whole compartmentalisation would not work)
-This also raises a question of what concentration of HCO3− can be transported within one vesicle, if the concentration is too low it will not function.
Transport of vesicles from bacteroid into the chloroplast: OMV
Could be largely based on the OMV-outer membrane transport, which has been worked out by igem team paris 2009. However a number of outer-transit/fusion peptides would have to be different to ensure targeting towards chloroplast and succesful fusion into the chloroplast.
problems:
-Usual transit peptide used for fusion protein targeting from cytoplasm into chloroplast (5kDa Rubisco subunit) might not work in targeting of the wholve vesicle into the chloroplast.
-Previous igem team have developed OMV to transport proteins from cytoplasm to another bacteria. In this situation however we would use OMV to transport concentrated solution from carboxysome - "organelle", therefore the OMV itself might not work on our setup.
Ideal solution: Engineer carboxysome with Carbonic anhydrase within plants (possibly within chloroplast) and use it to generate high CO2 concentration.
References: Moroney, J.V. & Somanchi, A., (1999). How Do Algae Concentrate CO2 to Increase the Efficiency of Photosynthetic Carbon Fixation? Plant Physiology, 119 (1), 9 -16.
Goodsell a S. Dutta, “Carbonic Anhydrase”, RCSB Protein Data Bank (january, 2004), http://www.pdb.org/pdb/101/motm.do?momID=49.
Nod factor interaction picture taken from: http://www.glycoforum.gr.jp/science/word/saccharide/SA-A02E.html (Text by Nick)
Food fermentation (food waste conversion/increasing shelf life)
- Thermoanaerobacterium thermosaccharolyticum can be used to convert food waste into hydrogen (1) However, I could not find information on the genes responsible for this and they may not have been identified yet.- Lactic acid bacteria can be used in food fermentation as "starter cultures". They produce several compounds and help extend the shelf life of the product (2).
- Lactic acid bacteria can produce a compound that fights Staph aureus, increasing food safety (3)
References:
(1) O-Thong, S., Prasertsan, P., Karakashev, D., & Angelidaki, I. (2008). Thermophilic fermentative hydrogen production by the newly isolated thermoanaerobacterium thermosaccharolyticum PSU-2. International Journal of Hydrogen Energy, 33(4), 1204-1214. and Shin, H. S., & Youn, J. H. (2005). Conversion of food waste into hydrogen by thermophilic acidogenesis. Biodegradation, 16(1), 33-44.
(2) Leroy, F., & De Vuyst, L. (2004). Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends in Food Science & Technology, 15(2), 67-78. (3) Cavadini, C., Hertel, C., & Hammes, W. P. (1998). Application of lysostaphin-producing lactobacilli to control staphylococcal food poisoning in meat products. Journal of Food Protection 174;, 61(4), 419-424. (Text by Rebekka)
Transportable Dengue Mosquitoes “simulated Nepenthes”
Engineering bacteria which could be put into the water container in order to attract, trap and kill the dengue mosquitoes and therefore decoy the mosquito away from biting you.Overview
- Dengue virus (RNA) : fever, haemorrhage, shock, organ disfunction
- Problems : no vaccine, difficult to engineer vaccine, acute but at the same time chronic
- Vectors : Aedes aegypti >> target!!!
- Why? : Lay egg at night (specific time), fragile, is the only 1 type of vector
Engineering bacteria
Module 1 : Allurement
- Odour > lactic a, octenol, NH3 > LDHB, LOX, AOS, GDH http://en.wikipedia.org/wiki/Aedes_aegypti, http://chemse.oxfordjournals.org/content/30/2/145.full
- Light > orange fluprescent protein (600 nm)http://journals.fcla.edu/flaent/article/view/75460
Module 2 : Entrapment
- Surfactant > Flavobacterium > GLD, https://colloque.inra.fr/flavobacterium/content/download/.../28Hunnicut.pdf
Module 3 : Torment
- Egg and Larva > Bacillus thuringiensis > Cry, Cyt, Chi
- Adult > Geranyl acetone > Populus trichocarpa > POPTRDRAFT_596199, http://www.ncbi.nlm.nih.gov/pubmed/20127888 (Text by Ming)
"DNA hard drive"
1. use DNA sequence as a medium to store information (DNA base - quaternary bit - original text)2. put “tag in” information (i.e some specific base pairs as “prototypes”) to classify the information
3. writing functions in programming language with DNA sequences, use DNA as the function variables
- Sci-Fi ideas developed with RCA people:
a. in the future, a bacteria charm/necklace can be made to store and carry the information, such as exam syllabus, genetic disease history, family photos, etc
b. different coding methods and types of bacteria can be chosen by the clients for different levels of security needs ,(some high risky information can be store in Bacillus anthraci for military use)
c. data can also be stored in E.coli in the human digestive system, it can be erased by intaking antibiotics
- practical problem: we need a bacteria platform to carry out the operation of the data (I.e: bio-compiler? bio-CPU?). A compiler consists of sets of programs and logic relations, it is theoretically workable, but obviously too difficult to do as a ten-week project
- since the bio-compiler idea is abandoned, we thought about other ways to store data:
- ROM memory: 8x8 grid to store 64 bits of data, the bacteria interaction can be modified to give to outputs, which indicates the two binary states (0 or 1)
- JK flip-flop or synchronized counter
- 7-segment display: one bacterium -four input channels-each channel with two states (0 or 1)-nine outputs to give nine numbers (0 to 9)- control the corresponding segments to display the number
-bacterial minesweeper. an inhibitor can be considered as a ”mine”. The main problem is that secondary diffusion is very difficult to control. a mass transfer equation must be modeled for each individual square
Bacteria that solidify the soil in the presence of urea
Sporosarcina pasteurii or Bacillus pasteurii from older taxonomies is a bacteria with the ability to solidify sand given a calcium and an organic nitrogen source through the process of biological cementation. This will be a good recycle of land waste, urea waste and a food waste.However solidification requires a high pH and produce toxic ammonium waste. Even though ammonia increase the pH this should be control using synthetic biology to model the right amount. ammonium can be subjected to other products which we are still searching for. Another application might be using ammonium produce to tighten the dye we made using the pigment which might allow the full house to be made easily from the brick. (Text by Ming)
Bile Acid Sensor
- the idea came up with Si’s final year project- a biosensor can be produced for daily home use to detect the bile acid concentration level in blood to prevent a series of liver diseases, especially the Intrahepatic cholestasis of pregnancy (ICP)
- the basic concept behind this sensor: bile acid - enzyme binding with the acid molecules – promoter – triggering the FXR gene – production of GFP
- GFP is normally used as an indicator of the bio-sensor. The main problem is that the fluorescent intensity is very hard to quantified for a home-user
- being inspired by the cosmetic skin colour sample card , we can make a sample card of fluorescence to give the rough concentration level of bile acid
- a threshold value is required to tell the patient when their blood bile acid level is dangerous and may need a medical treatment
- therefore, we will modify the linear relationship between bile acid concentration and GFP intensity level into a Hill system using Hill equation to find the threshold value
- also, we may use colour indicator instead of GFP (light indicator)
Background:
- Bile acids are 24-carbon steroids found in bile, which are subject to enterohepatic circulation
- synthesized in liver and stored in gallbladder, helping in digestion and absorption of dietary fat and liposoluble vitamins
- bile acids are highly toxic. Therefore their concentration must be tightly regulated
- the level of bile acids is controlled through a negative feedback system mediated by a nuclear bile acid receptor FXR. FXR is highly expressed in liver, intestine and kidney cells. It responds to bile acids and has been shown to repress CYP7A1, a key gene associated with bile acid synthesis.
- Defects in bile acid homeostasis due to functional variations of FXR result in cholestatic conditions such as Intrahepatic cholestasis of pregnancy (ICP). ICP is a pregnancy-specific liver disorder characterized by pruritus (intense itch) an abnormal liver function
- The dysfunction of maternal liver could induce stress on the fetal liver as the fetus relies on maternal liver to remove bile. It has been shown that ICP pregnancies are more likely to suffer from meconium staining of the amniotic fluid (MSAF), cardiotocography (GCT) abnormalities and respiration distress syndrome (RDS).
- The risk of complications such preterm labor, prenatal death and stillbirth are directly linked to severity of ICP
- The morbidity of ICP is geographically and racially dependent. [1]
Country Morbidity(%)
Sweden 4.2
Finland 1.0
Poland 1.5
Jugoslavia 1.1
Spain 1.6
UK 1.0
China 4.4
Aymara 13.8
Araucanian 27.6
Chile 15.6
Caucasian species 9.8
Mechanism:
FXR based activation:
- use eukaryotic transcription factors in bacterial gene expression
- suggested use of FXR (known structure and DNA sequence) with constitutive promoter expression
- a receptor protein of a number of bile acids, which would bind to ligand-bile acid
- can act as a repressor or an activator, we would use it as an activator, binding to a promoter region B4-BARE (2.4kb) taken from a gene UGT2B4, (known primers for promoter region)
- specified exact FXR binding site, therefore possibility of integrating FXR binding site into another regulatory region
- upon binding to bile acid it would activate output gene
- In the experiment using FXR as transcription factor in human cells 30mM DCA was used, however presumably lower concentration should be sufficient for triggering of FXR.
Output:
- GFP: fluorescence = light indicator = hard to quantify
- ligaments: colour = clear indicator = influenced by the red colour of blood cells = cell free mechanism for hospital use = accurate measurement = filter/ membrane mechanism for home use
- protein expressed on surface of bacteria causing aggregation (either of bacteria or of bacteria to a substance in blood)
Modelling:
- Binding of activator to GFP gene is a positive cooperative reaction.
- Once activator molecule is bound to the enzyme, its affinity for other activator molecules increases.
- Hence, Hill equation can be used as a model.
Hill function for transcriptional activation:
k1: Maximal transcription rate
Km: Activation coefficient
n: Hill coefficient
A: [activator]
- This equation gives the % bound by activator as a function of activator concentration.
- after Hill equation modification, the system behaves like a switch
- In addition, we also need to model the diffusion of blood and GFP in the bacteria culture
Cell –free mechanism:
- for hospital use, the blood must be processed through a cell-free system to give an accurate test result, as well as the bacteria to reduce the risk if they leak from the container ( after the cell-free process, the bacteria are not able to reproduce)
- separate certain organelles from whole cells for further analysis of specific parts of cells
- in the process, a tissue sample is first homogenised to break the cell membranes and mix up the cell contents
- homogenization is intensive blending of mutually related substances or groups of mutually related substances to form a constant of different insoluble phases
- homogenate is then subjected to repeated centrifugations
- each time removing the pellet and increasing the centrifugal force
- Separation is based on size and density, with larger and denser particles pelleting at lower centrifugal forces. In the separating order in actual application: Whole cells and nuclei; Mitochondria, lysosomes and peroxisomes; Microsomes (vesicles of disrupted endoplasmic reticulum); ribosomes and cytosol.
Filter/semi-permeable membrane for the blood cells:
RBC: 7~8.5μm
neutrophil: 10~12μm
eosinophil: 10~15μm
basophil: 10~12μm
monocyte: 14~20μm
lymphocyte: small: 6~8μm, medium9~12μm, large: 13~20μm
taurocholic acid molecule roughly 0.4nm
- therefore, if the hole diameter of the filter is set to be at about 2~5nm, all the blood cells can be filtered
Human practices (with help of LSE BIOS):
A. why biosensors ?
- possible to construct
- has a potential market
- B. human practices
1. bio-safety
- cell-free system to stop the bacteria from reproducing if there is a leakage of the sensor device
- (unpredictable) mutation must be carefully prevented during the engineering part
2. bio-security
- the access to DNA sequences and other genetic information must be controlled
- “garage biologist”
- the restriction to synthetic biology knowledge is not the way to prevent bio-terrorism, the key thing is the professional and correct guidance
3. IP (intellectual property issue) and patent
4. ethical and philosophy
5. global fairness
- bio-sensing = faster disease detection
- employment problem? discrimination?
- classify the diseases into different levels of risks
- social coordinate organization to optimize the occupation and personnel resources (Text by Si, Nina, Nick and Yuanwei)
Inflammation detecting bandage
- inflammation is caused by the immune response to pathogens- can be acute to chronic
- acute inflammation:
1. increased movement of plasma and leukocytes (granulocytes) from blood to injured tissues
2. causative agent = pathogens and injured tissues
3. cells involved: neutrophils, mononuclear cells (monocytes and macrophagens)
4. primary mediator = vasoactive amine and eicosanoids
- to carry out the detection mechanism, we will set a target chemical to detect
- at this stage, interferon gamma is taken into our consideration: Interferon-gamma (IFN-γ) is a dimerized soluble cytokine that is the only member of the type II class of interferon. This interferon was originally called macrophage-activating factor, a term now used to describe a larger family of proteins to which IFN-γ belongs. In humans, the IFN-γ protein is encoded by the IFNG gene.[2]
- the goal of this project is to find a gene coding the protein which and react with interferon gamma and give an indication
- a further step may be taken as a “damaged tissue cleaner”, which means that the bacteria can not only detect but also remove the inflammation tissue
[1] http://baike.baidu.com/view/676699.htm
[2] http://en.wikipedia.org/wiki/Interferon-gamma (Text by Si, Nina, Nick and Yuanwei)
Sand brick produced by bacteria
Sporosarcina pasteurii --> ureaseCalcium carbonate bricks can be made by using urease, urea, sand and calcium chloride. The urease cleaves the urea and creates ammonia and carbon dioxide. The ammonia raises the pH and causes the calcium ions to precipitate with the dissolved carbonate ions.
Ammonia is currently a problem since it can run off to cause algal bloom. A method to remove ammonia must be obtained. Solution? Possibly express all the urea cycle enzymes in the chassis (5 total enzymes). Use slovenia's DNA scaffold so that the process does not produce too many possibly problematic intermediaries in the bacteria.
When should ammonia be converted back to urea? When calcium is depleted. Need some sort of Calcium sensor.
http://www.msnbc.msn.com/id/32558231/ns/technology_and_science-science/t/study-bacteria-can-make-salt-water-drinkable/
So that's pretty cool, but perhaps the bacteria that store salt can be used to spread salt in the winter and stop the country grinding to a halt every time it snows? (Text by Frank)
New way of transforming bacteria idea
Create a plasmid containing Dpn1 under the influence of a strong repressor. In the same plasmid insert the methylase enzyme. The gene is inserted into the methylase and stops its transcription.If the gene of interest has been successfully inserted into the plasmid and the plasmid has been successfully inserted into the bacteria we can use the inducer to kill off any bacteria that has not been transformed (because the Dpn1 cuts any methylated strands of DNA).
Today we continued developing the bile acid sensor as well as the idea of the anti-venom. For the anti-venom idea we talked to Travis Bayer and obtained a method for screening for the right mutants. We also had a visit from LSE BIOS members that taught us about the point points we should worry about in human practices. We were also given the task to make instructions for a lego structure to show us how difficult it is to standardize parts due to the difficulty of communicating more complicated instructions. (Text by Chris)
Making rain with bacterial spores
The surface of spores can form crystal structures that attract water vapour from the air and form rain drops. We wanted to use this priniciple to have a useful output from the spores when they reach the ground - which could be sand or soil.There are several genes localized that contribute to the spore outer layer which could potentially be randomly mutated to optimize the geometric shape of the spore to promote water droplet formation.
One of the drawbacks to using spores is that we need germination to occur for gene expression and there is an obvious risk of releasing GMOs into nature. One option could be to engineer in a death response to have limited gene expression of mucin for example which could aid in water retention, or auxin to promote root growth deep into soil.
Greenhouse Gases (isoprene)
1.Water vapor2. Carbon dioxide - done to death
3. Methane
4. Ozone
CFC's, VOC's and Nitrous oxide. CFC's are banned. However, chlorine radicals still a problem.
isoprene is produced by plants to prevent oxidative stress and heat shock. VOC. Byproduct of the thermal cracking of naphtha or oil. Used to produce natural rubber...
Produced from the precursor DMAPP
DMAPP is made by isopentenyl pyrophosphate isomerase from IPP.
IPP is also one of the precursors of of lycopene. You need 3 IPP, 1 DMAPP and 1 GGPP (Geranylgeranyl pyrophosphate) to produce Phytoene.
Insert lycopene biosynthesis pathway and inhibit isoprene synthase?
Probably too complicated.
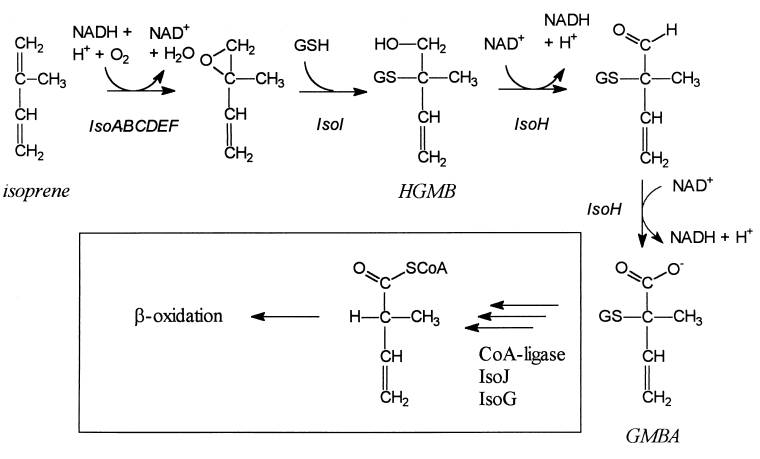
1. Johan E. T. van Hylckama Vlieg et al., “Characterization of the Gene Cluster Involved in Isoprene Metabolism in Rhodococcus sp. Strain AD45,” Journal of Bacteriology 182, no. 7 (April 2000): 1956-1963.
Rhodococcus sp. strain AD45
Isoprene is supposed to recycle OH. NOx causes oxidant build-up.
Mechanism for NOx removal?
NO sources: Internal combustion engine. Cannot target.
NO2 sources: Internal combustion engine, thermal power station, pulp mill. None of which could be targeted.
Pulp mill industry is one of the largest producers of water pollution in the world. An alternative to paper could solve a lot of problems. Still unfeasible.
Containment is an issue.
Estrogenic endocrine disruptors in waste water
- Estrogenic compounds are released by multiple sources (1)- In addition, many compounds in drinking water can act as mimics of estrogen in human and animal bodies (2)
- This is problematic because endocrine disruptors interfere with the hormone balances of humans and animals, in particular fish (9)
- Biosensors for this already exist (3), (4)
- This could perhaps be coupled to a degradation mechanism, involving e.g. laccases, which degrade one type of xenoestrogen (5)
- However, laccase degradation of bisphenol A leads to the production of corrosive compounds (6). It may therefore by preferable to use horseradish peroxidase, which appears to be able to degrade a range of estrogens and estrogen-like compounds (7)
- Horseradish peroxidase works most efficiently at around neutral pH (8), which is close to that of most waste water (buffering may be an issue?).
- This peroxidase is especially useful as its reaction products non-enzymatically aggregate and can be separated from waste water by traditional processes. Horseradish peroxidase also degrades phenol (7), which may give it another useful application in clearing industrial waste from waste water.
- Maybe modify chemotaxis to make bacteria swim towards target? Might be able to use the module from Imperial iGEM '08 (Text by Rebekka)
References:
(1) http://www.sciencedirect.com/science/article/pii/S0160412002000752
(2) http://pubs.acs.org/doi/full/10.1021/es801845a
(3) http://www.sciencedirect.com/science/article/pii/S0022283605004080
(4) http://www.sciencedirect.com/science/article/pii/S0048969709012303
(5) http://mic.sgmjournals.org/content/151/1/45.full
(6) http://www.sciencedirect.com/science/article/pii/S096085240500338X
(7) http://www.sciencedirect.com/science/article/pii/S0045653507004183
(8) http://www.sciencedirect.com/science/article/pii/S0043135406003228
(9) http://www.sciencedirect.com/science/article/pii/S0048969709005579
Bacteria targeting parasitic nematodes
Plant parasitic nematodes Plant parasitic nematodes (PPNs) cause billions of dollars in crop damage annually and the only relatively effective solution is chemical which cause environmental damage.Biological solutions: Nematode populations have been declined by biological microbes- mainly by two parasitic fungi, Nematophthora gynophila and Verticillium chlamydosporium, which attack the developing female on the root surface. 95 to 97 % of the females and eggs are destroyed (Kerry, Crump and Mullen, 1982). Thus the natural control of cereal-cyst nematode in a range of soils is predictable and effective, but slow acting.
See: http://www.fao.org/docrep/V9978E/v9978e0b.htm for a list of potential biological agents and their shortcomings
The two most problematic species, the root-knot and cyst nematodes, infect roots of plants and feed off of their tissue. Nematodes are known to find food by chemotaxis and one type of bacteria, Bacillus nematocida, attracts nematodes by secreting volatile organic compounds and is then ingested by the worms. When it adapts to the intestine, it secretes proteases to kill the nematode.
This principle could be applied to an engineered bacteria that secretes a compound to attract PPNs (it would have to attract only PPNs, because there are a lot of good nematodes in soil) and once ingested kill the nematodes by RNAi knockout of the parasitic genes or genes involved in reproduction. A problem with this is that PPNs eat plants, not bacteria.
Newly hatched larvae have to migrate through the soil efficiently to locate plants because they have limited nutrient resources. IAA (the main type of auxin) and other indole compound gradients have been shown to attract PPNs, allowing plant infection. As long as the RNAi target is specific to PPNs, attraction of the PPNs does not have to be as specific.
Successful RNAi PPN targets in GM crops are a gene encoding a splicing factor and a gene encoding an integrase to target the root knot nematode M incognita. Most of the genes expressed in the esophageal gland cells (generate molecules secreted into the plant through the stylet) of PPNs encode proteins secreted into the host root (include cell wall modifying enzymes, regulators of host cell cycle & metabolism, suppressors of host defence, and mimics of plant molecules)
Problem: Can we get relatively specific chemo-attraction of PPNs? How do we introduce RNAi into the nematodes? How do we contain the GM bacteria in a field and will PPNs be able to migrate effectively to these contained areas?
Lymphatic filariasis
Second leading cause of longterm disability worldwide (as a result of lymphedema, elephantiasis, hydrocele and periodic fevers), is caused by mosquito-transmitted filarial worms, including Wuchereria bancrofti and Brugia malayi, which colonize the lymphatic system. Drug treatments are fairly effective at killing larval stages but don’t provide much benefit to infected hosts
Research is being done to determine important genes in the nematodes via RNAi.
How could we target this nematode to prevent mosquito infection and subsequent human infection?
Termites
Termites are known for their ability to digest nutritionally poor food sources such as lignocellulose. A recent paper has studied the synergistic properties of cellulases from Reticulitermes flavipes. Three enzymes were studied, two cellulases and a laccase. The cellulases were 1 β-glucosidase and Cell-1 (a GHF9 endoglucanase). The phenol-oxidizing laccase that is involved in lignin degradation is LacA.Cell-1 and β-glu showed >300-fold and >70-fold increases in glucose production (when degrading pine sawdust and beechwood xylan) when combined.
When Lac-6 was added in the pine saw-dust experiment, glucose production rate decreased. The authors say that this is likely due to the process of end-product inhibition on β-glu.
However, glucose production increased when all three enzymes were used on beechwood xylan. Lac-6 enhances glucose release from hemicellulose by cellulases?
Microcrystalline cellulose results were nearly identical to sawdust.
The idea of this project is to use bacteria to express Lac-6, β-glu and Cell-1 in order to produce monosaccharides from other sources of biomass such as wood. It has recently been discovered that these termite derived cellulases have synergetic properties. Therefore I postulate that future effective biomass conversion will rely on obtaining the most efficient mixtures of enzymes.
Lignin (the material hardest to digest) releases pentoses (like xylose and arabinose). Since sawdust degradation by the three enzymes works best when only Cell-1 and β-glu are used whereas all three enzymes gave the largest production rate when degrading beechwood xylan, I propose the construction of a module that will allow the bacteria to produce the most efficient enzyme mixtures depending on the rate of glucose production. This circuit might make it possible to digest complex mixtures of materials most efficiently. The bacteria used should also ideally only use an alternative carbon source to glucose.
Using anammox bacteria to treat eutrophication:-
- eutrophication = biomass accumulation in waterbodies- causes unbalanced spieces
- anammox bacteria = anaerobic ammonium oxidation
NH4+ + NO2- = N2 + H2O
turning ammonium and nitrite into di-nitrogen gas
- Anammoxosome
special membrane bound compartment
consists of ladderane lipids
• Consist of 2 or more cyclobutane rings
• Highly strained structure
• Can be isolated from ammonia oxidising bacteria living in sea
• Process of synthesis not clear
• Anaerobic ammonium oxidising bacteria have cell membranes consisting of concatenated cyclobutane moieties (ladderane lipids). This structure is a result of the process of converting ammonia to nitrogen gas in the absence of oxygen.
• One possible pathway: Gene clusters of K. stuttgartiensis -> anaerobic synthesis of PUFA -> ladderane lipids.
• Possible application as building blocks in optoelectronic
Anammoxosome contains the enzymes of the redox reactions, prevents the poison intermediates (hydrazine and hydroxylamine ) from killing the bacteria.
Reaction process:-
- Reduction of nitrite by nitrite reductase
- Production of hydrazine by hydrazine hydrolase
- Oxidation of hydrazine into nitrogen gas by oxidizing enzyme
- Production of ATP (proton motive force)
Enzyme sulfite reductase (ferredoxin):-
ammonium + oxidized ferredoxin + H2O = nitrite + reduced ferredoxin + H+
ferredoxin acts as an electron transfer of this reaction
may this reaction bi-pass the production of hydrazine ?
Polycyclic Aromatic Hydrocarbon (PAH)
Problem: PAHs are a class of organic compounds that are toxic and carcinogenic. PAHs are released into the environment through processes like the combustion of fossil fuels, plastic, coke production and vehicle exhaust. PAHs can be found in air (cigarette smoke) and accumulate in soil. It can also be found in water due to industrial effluents and oil spills. The poor water solubility of PAH causes it to be deposited in sediments and aquatic organisms, affecting fish farms, etc.
Solution: Factories release PAHs into water or ponds which affect survival of fishes in fish farm. Bacillus subtilis can break up the PAHs and eliminate this pollutant. In addition, the Bacillus also contains gene which codes for the production of vitamin which is beneficial to the growth of the fish. To exploit this benefit, we need to find a way for Zooplankton (found in aquatic environment) to incorporate the Bacillus and its vitamin-producing gene. The fish feeding on the zooplankton will then receive a good source of vitamin for its growth. To do this, we can engineer Bacillus bacteria to act as a vector which can deliver vitamin to the cytoplasms of zooplankton which is a phagocytic eukaryote. Essentially, this project allows us to not only clear up the pollutant PAH, but it also provides a source of nutrients for the fish. In addition, the waste product of the fish can also be used by the Bacillus to provide nutrients for the fish.
Filling in the green gap
-plants absorb light wavelength from 400-500nm and then 600-700nm however between 500 - 600nm, plants do not have any light accepting compounds in that light range
-there are two possible ways of solving the problem:
1.introduce lightharvesting pigments from red algae and couple them to plant photosystems
2.use any fluorophore which accepts light in the green gap and emits the light in wavelength higher than 600nm, localise it on the membrane on chloroplast and therefore transfer wavelength between 500-600nm to a wavelength higher than 600nm so plant phytochromes can accept it
-it appears that red algae combine the two methods in phycobilisomes, where they use pigments which emit wavelengths a bit higher than the emmited wavelength until chlorophyll is capable of accepting light
-possbility of introducing phycobilisomes or only specific phycobiliproteins, which accept (500-600nm) and re-emit it in 600-700nm
-problem: does not seem to be a single protein for accepting and re-emitting of the corresponding wavelengths
-problem: do not know the number of phycobiliproteins neccessary to significantly contribute towards excitation of plant phytochromes
-problem: plant might be losing energy by synthesising extra pigment proteins (if it needs a lot of them) even though these proteins would contribute to produce more energy in the plant
Auxin-Secreting Bacteria
The project intends to enhance the ability of plants to grow roots towards specific locations. For example, this could be used to allow plants to quickly locate the nutrients they need with minimal energy expenditure. Another example is to increase the speed at which roots grow, in order to hold down soil that is in danger of being eroded.
This would be done using a two bacteria system, so that one strain of bacteria would associate closely with the roots of a specific plant and another strain could locate the target and secrete a long-distance signalling molecule that would cause the root-associated bacteria to secrete auxin, causing the roots to grow.
To model this, a simple system will be used, using Azotobacter paspali, which associates specifically with the roots of paspalum grass. The bacteria would be modified to secrete auxin in a dose-dependent fashion in response to a signal. The exploratory strain of bacteria would be engineered to seek out specific conditions and to release a signalling molecule.
The signalling molecule could be glutamate, calcium, possibly a homoserine lactone in a vesicle, Paris 2009.
In this paper, http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0011915 , it was shown that tomatoes and arabidopsis plants are able to take up microbes to degrade them. The bacteria were alive for around 10 days before they were all completely gone. It might be possible to have these bacteria secrete nitrates and auxin once inside the cell, or over-express it so that it is released when the bacteria are lysed by the plant.
The bacteria could potentially be engineered to thrive inside the plant roots. For this, it is important to note that it is the plants that break down their own cell walls and actively take up the bacteria.
Could also use yeast.
Converting PAH to PHA
PAH or poly aromatic hydrocarbon has been one of the major water pollutant in many tropical countries due to incorrect water waste regulation in the plastics and fuel industry. Its carcinogenicity is considered highly toxic to both human and the river creatures and hence causes major health and agricultural problem. PAH primarily accumulates inside the soil and contaminates the plankton on the riverbed. The toxicity is transmitted as the fish eat the plankton and is eaten by human respectively. Our idea is to convert this highly dangerous substance into a useful product where PHA ,polyhydroxyalkaonate, since it uses the same precursor that could be broken down by PAH, acetyl CoA and succinyl CoA. PHA is also biodegradable, thereby safe for environment, has higher elasticity, strength and and thermal stability than polyethelene and polypropylene, the non-biodegradable plastics using in everyday-life.
The solution is by engineering the bacteria that can convert the PAH to acetylCoA using the mechanism below. The bacteria can sense the breakdown of catechol which will acts as a transcription activator for the expression of AHL, allows the aggregation of the bacteria to promote PAH breakdown into acetyl CoA and sucinyl CoA. Another module that is incorporated into the bacteria are the genes responsible for PHA production as well as the gas vesicle protein and RFP. Therefore after collecting the plastics (density = 1.48 g/ml > water density) The bacteria can float up and will be easily detected. Eventually the plastic producing bacteria could be collected for plastics extraction and the region contaminated by PAH could be identified where more bacteria could be applied for more breakdown.
 "
"




