Team:HokkaidoU Japan/Project/Onion
From 2011.igem.org
m |
|||
| (2 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
==SUMMARY== | ==SUMMARY== | ||
| - | Last year, we have achieved | + | Last year, we have achieved injection of GFP to the RK13 cells (cultivated rabbit kidney cell) using ''E. coli'' expressing T3SS. However, using animal cells is inconvenient. For example, cultivation of the cells a must be started couple of days before infection. And deterioration of cultivation environment results in immediate death of cells. We thought that we might solve these problems by using onion cells. Onion has very widely available and its cells are tolerant of bacteria. Moreover, since onion forms detachable monolayer cell-sheets,they can be observe with ease. We repeatedly tried to establish injection protocol for onion cells and after some trials we were successful. |
==MATERIALS== | ==MATERIALS== | ||
| - | * ''E. coli'' strain | + | * ''E. coli'' strain which can inject GFP into eukaryotic cells that we found last year. |
* Epidermal cell sheet of onion | * Epidermal cell sheet of onion | ||
* Reagents | * Reagents | ||
| Line 23: | Line 23: | ||
==PROCEDURE== | ==PROCEDURE== | ||
===Prepararion of bacterial suspension=== | ===Prepararion of bacterial suspension=== | ||
| - | # Isolate a single colony of ''E. coli'' and cultured cells in liquid LB ( | + | # Isolate a single colony of ''E. coli'' and cultured cells in liquid LB (without antibiotics) for 2 hours. |
| - | # Add 500 ul of culture fluid to 2 ml of liquid LB. Add appropriate amount of antibiotics (Tetracycline and Chloramphenicol). Add 50ul of 20% L-arabinose solution. | + | # Add 500 ul of culture fluid to 2 ml of liquid LB. Add appropriate amount of antibiotics (Tetracycline and Chloramphenicol). |
| - | # Culture cells overnight at 37C, 200rpm | + | # Add 50ul of 20% L-arabinose solution. |
| - | # | + | # Culture cells overnight at 37C, shaking at 200rpm |
| - | # Add MgM-MES (pH5.0), Tetracycline, Chloramphenicol, L-arabinose to the bacterial pellet. Resuspend it using vortex mixer. Culture cells at 37C, 200rpm | + | # Spin down the cultured cells at 3000rpm, for 10 min at 35C. Remove supernatant. |
| - | # | + | # Add MgM-MES (pH5.0), Tetracycline, Chloramphenicol, L-arabinose to the bacterial pellet. Resuspend it using vortex mixer. Culture cells at 37C, shaking at 200rpm for 4 hours. This step is required because genes encoding salmonella T3SS are expressed in acidic environment. |
| - | # Resuspend the pellet with 2 ml of MgM-MES (pH5.0) containing 0.4 M Mannitol, then | + | # Spin down the culture fluid at 3000rpm, for 10 min at 25C. Remove supernatant. |
| + | # Resuspend the pellet with 2 ml of MgM-MES (pH5.0) containing 0.4 M Mannitol, then spin down the culture fluid at 3000rpm, for 10 min at 25C. Remove supernatant. Repeat this step 3 times. (This is done to remove the toxic substances that are produced by ''E. coli'' and adjust the osmotic pressure to that in onion cells) | ||
# Resuspend the pellet with 2 ml of MgM-MES (pH5.0) containing 0.4 M Mannitol, Tetracycline, chloramphenicol and L-arabinose. | # Resuspend the pellet with 2 ml of MgM-MES (pH5.0) containing 0.4 M Mannitol, Tetracycline, chloramphenicol and L-arabinose. | ||
| - | # Measure the absorbance at 600 nm using a spectrophotometer, then adjust the concentration of cells to | + | # Measure the light absorbance at 600 nm using a spectrophotometer, then adjust the concentration of cells to ΔOD600 = 0.06 by diluting with the same medium mentioned above. |
| - | # | + | # To infect onion cells with ''E. coli''add 500 ul of the culture fluid onto pre-treated onion cell-sheets. Leave at RT in petri dish (Prevent drying) |
| - | # Remove the bacterial culture fluid carefully by micropipette so that the cell-sheets are not to be torn. Observe the cells under a | + | # Remove the bacterial culture fluid carefully by micropipette so that the cell-sheets are not to be torn. Observe the cells under a fluorescencent microscope. |
===Preparation of onion cells=== | ===Preparation of onion cells=== | ||
| + | [[File:HokkaidoU Onion Procedure.png|100px|right|]] | ||
Since we thought that the cell wall may prevent injection of protein, onion cell-sheets were pretreated to remove cell wall as follows. | Since we thought that the cell wall may prevent injection of protein, onion cell-sheets were pretreated to remove cell wall as follows. | ||
| - | # Cut | + | # Cut an onion into four pieces. |
# Take out a fresh sheets of epidermal cells (second or third layer from the surface of onion). | # Take out a fresh sheets of epidermal cells (second or third layer from the surface of onion). | ||
| - | # Cut the layer into rectangular (about 1.5cm x 3cm) | + | # Cut the layer into rectangular shape (about 1.5cm x 3cm) |
| - | # Tear off a cell sheet from the inner | + | # Tear off a cell sheet from the inner surface of the layer. |
| - | + | # Put the sheet onto a slide glass at back face up because the upper face deflects enzyme solution, and fix the edge of the sheet onto slide glass with 1% agarose gel. | |
| - | # Fill a petri dish with 1% cellulase and 0.1% | + | # Fill a petri dish with 1% cellulase and 0.1% pectolyase in 0.4 M Mannitol solution (pH7.0), then submerge the cell sheet fixed on slide glass into the enzyme solution. Incubate for 8 min at RT. |
| - | + | # Prepare 3 petri dishes filled with MgM-MES pH5.0 containing 0.4 M Mannitol. After processing, wash cells stepwise soaking in these petri dishes to remove enzyme solution. | |
| - | # Prepare 3 petri dishes | + | # Remove excess moisture, and transfer the sample to another petri dishs for infection. |
| - | # Remove excess moisture, and transfer the sample to another petri dishs for infection. | + | <div style="clear:both;"></div> |
| - | + | ||
| - | + | ||
| - | + | ||
==Results== | ==Results== | ||
We carried out infection assay using following ''E. coli'': | We carried out infection assay using following ''E. coli'': | ||
| Line 56: | Line 55: | ||
# T3SS only | # T3SS only | ||
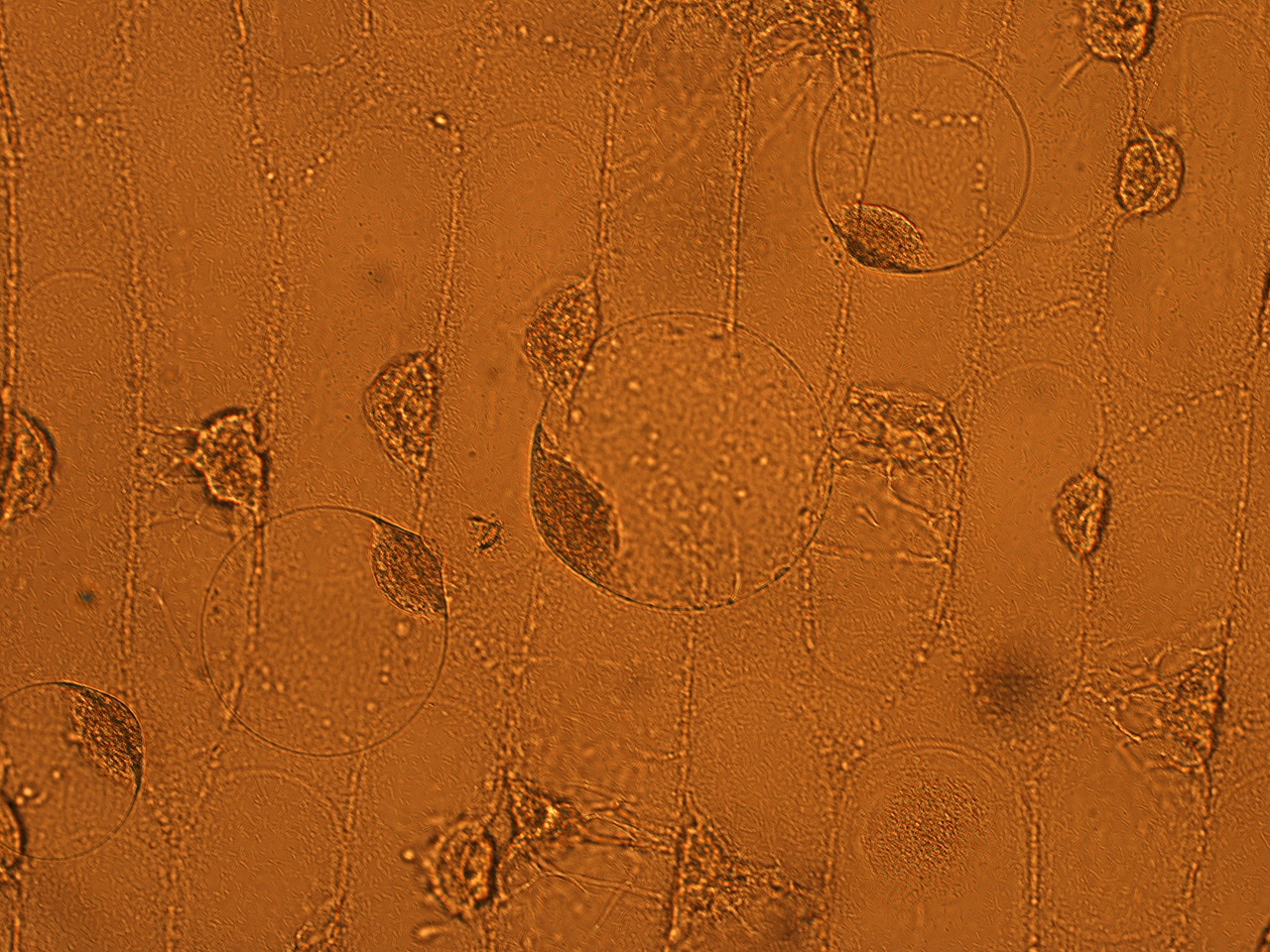
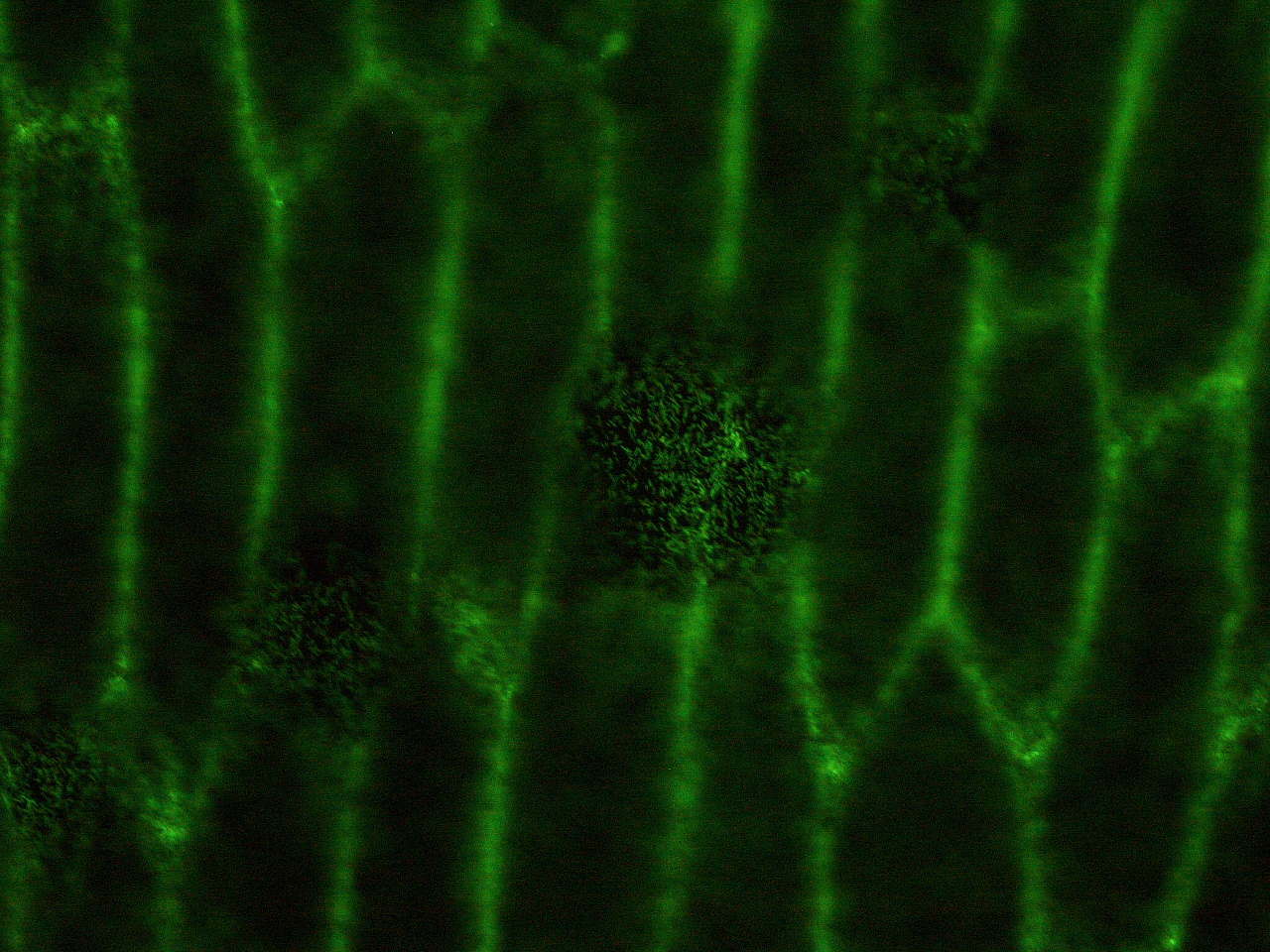
| - | Here | + | Here are the results of infection assay figure 1 to 7. Figure 1 to 3 shows the results of infection assay using the ''E. coli'' which we have made last year. That ''E. coli'' has T3SS, GFP with secretion signal, and RFP. |
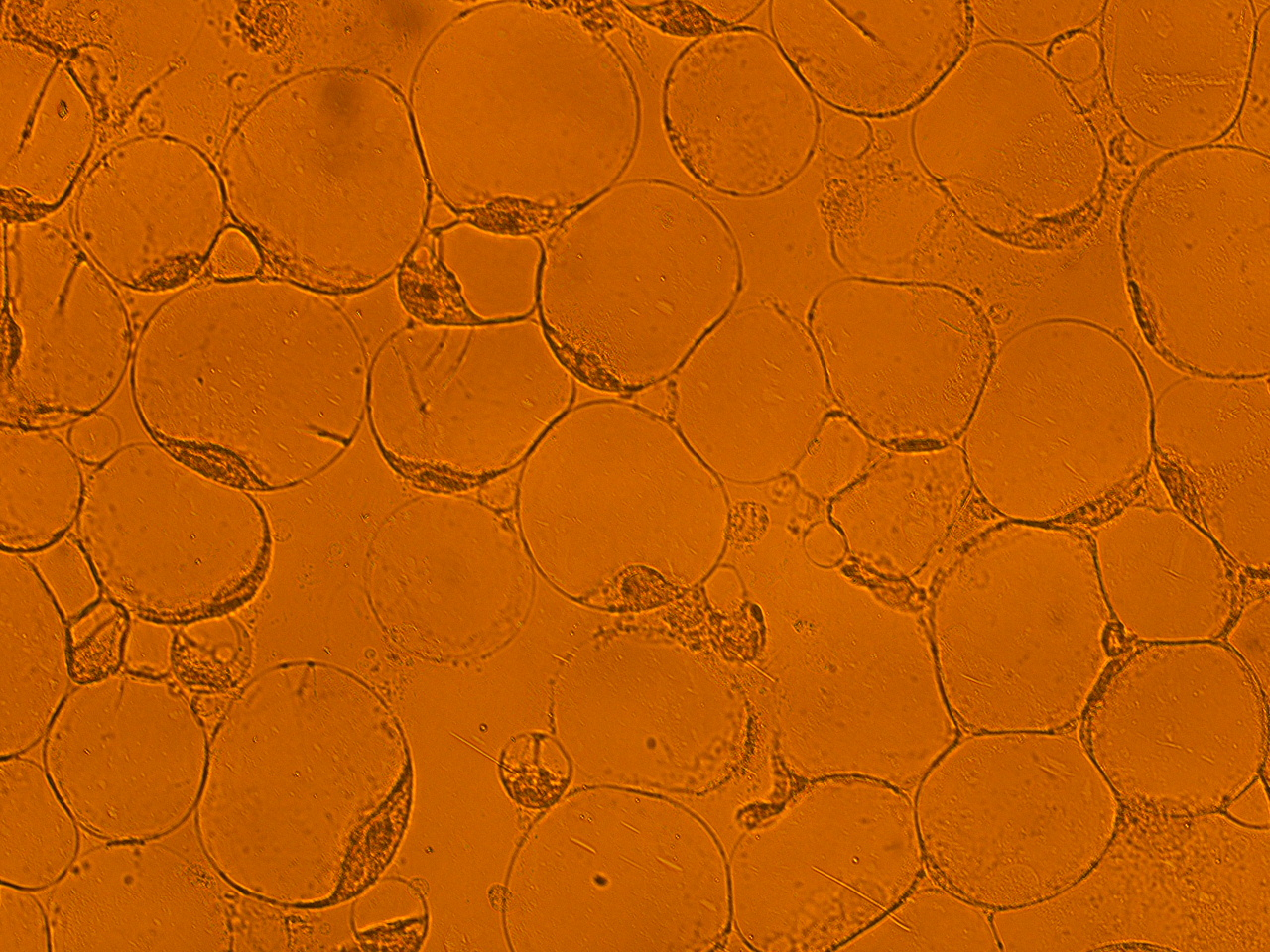
[[File:HokkaidoU_onion_1.JPG|thumb|300px|left|Fig. 1 Onion cells after injection was taken under bright field.]] | [[File:HokkaidoU_onion_1.JPG|thumb|300px|left|Fig. 1 Onion cells after injection was taken under bright field.]] | ||
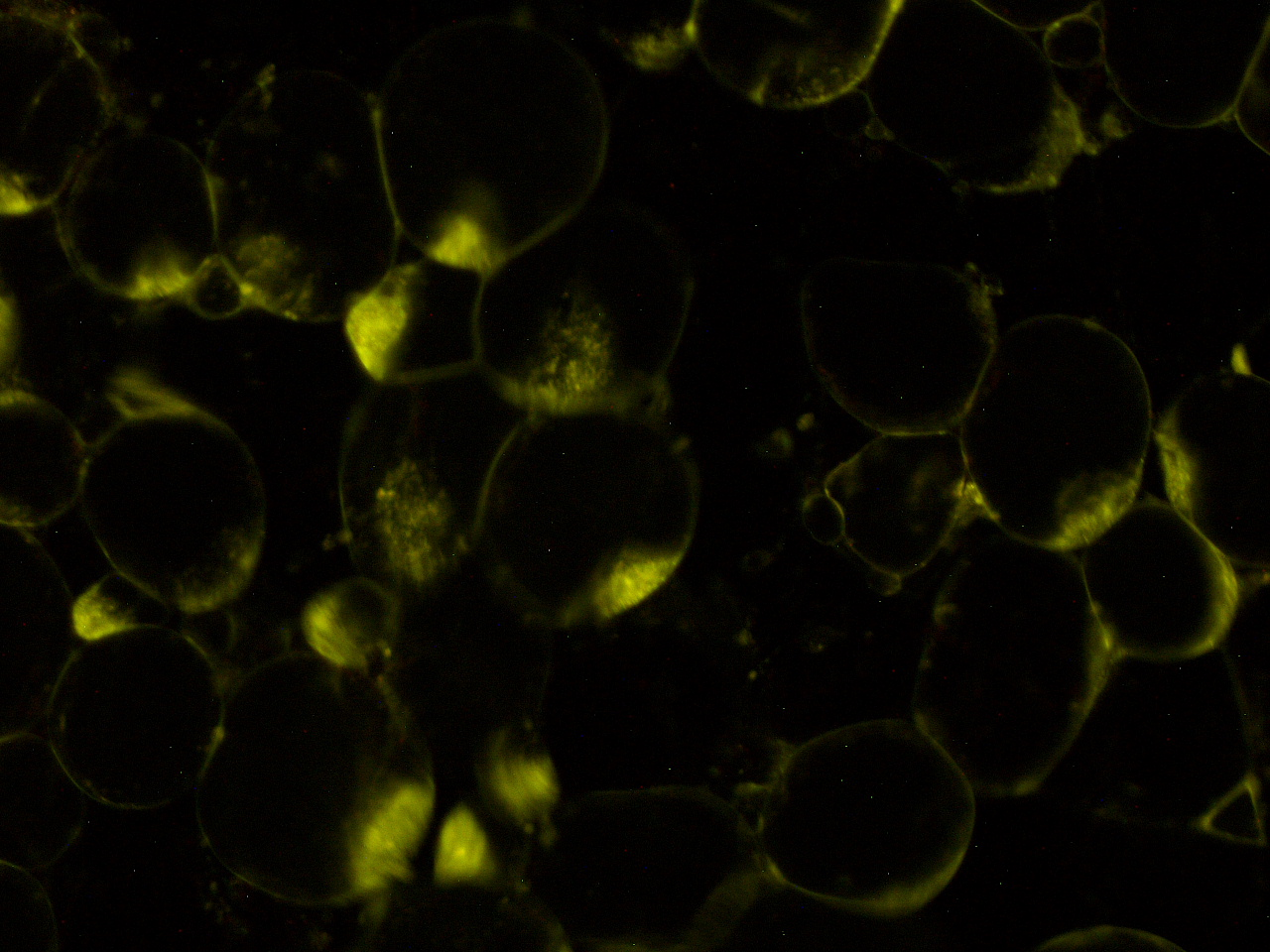
[[File:HokkaidoU_onion_2.JPG|thumb|300px|left|Fig. 2 Subcellular localization of GFP after injection. The picture of onion cells was taken under blue light to observe GFP that is fused to secretion signal.]] | [[File:HokkaidoU_onion_2.JPG|thumb|300px|left|Fig. 2 Subcellular localization of GFP after injection. The picture of onion cells was taken under blue light to observe GFP that is fused to secretion signal.]] | ||
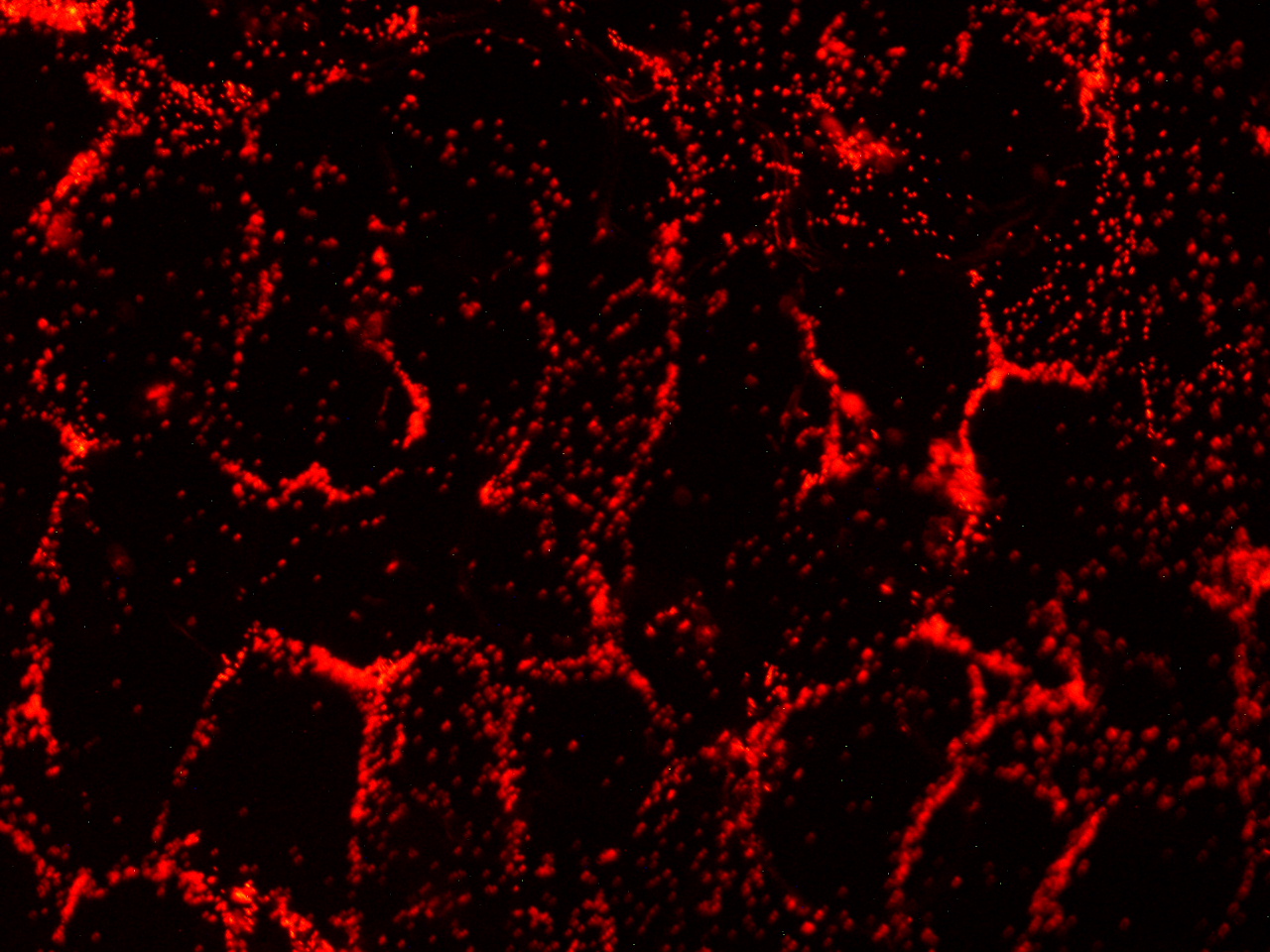
| - | [[File:HokkaidoU_onion_3.JPG|thumb|300px|left|Fig. 3 | + | [[File:HokkaidoU_onion_3.JPG|thumb|300px|left|Fig. 3 RFP remains in ''E. coli'' . The picture was taken under green light to observe RFP that is not fused to secretion signal.]] |
[[File:HokkaidoU_onion_4.JPG|thumb|300px|left|Fig. 4 Onion cells after injection was taken under bright field.]] | [[File:HokkaidoU_onion_4.JPG|thumb|300px|left|Fig. 4 Onion cells after injection was taken under bright field.]] | ||
[[File:HokkaidoU_onion_5b.JPG|thumb|300px|left|Fig. 5 Localization of GFP signal associated with ''E. coli'' cells. The picture of onion cells was taken under blue light to observe GFP that is fused to secretion signal.]]<div style="clear:both;"></div> | [[File:HokkaidoU_onion_5b.JPG|thumb|300px|left|Fig. 5 Localization of GFP signal associated with ''E. coli'' cells. The picture of onion cells was taken under blue light to observe GFP that is fused to secretion signal.]]<div style="clear:both;"></div> | ||
| - | GFP not fused to secretion signal is not injected to the onion cells. | + | |
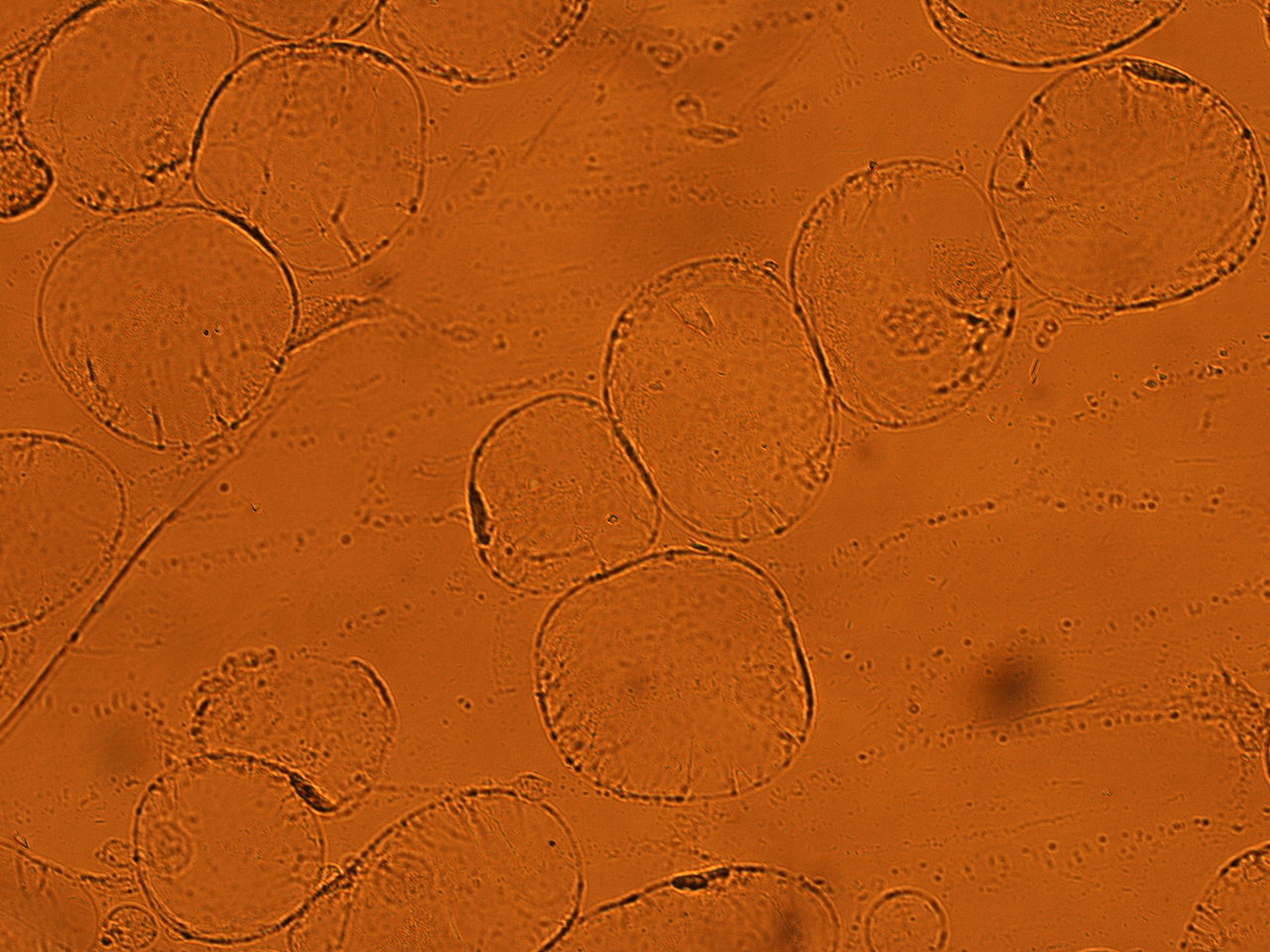
| + | As shown in following pictures GFP not fused to secretion signal is not injected to the onion cells. | ||
[[File:HokkaidoU_onion_6.JPG|thumb|300px|left|Fig. 6 The picture of onion cells after injection was taken under bright field.]] | [[File:HokkaidoU_onion_6.JPG|thumb|300px|left|Fig. 6 The picture of onion cells after injection was taken under bright field.]] | ||
| Line 70: | Line 70: | ||
<div style="clear:both;"></div> | <div style="clear:both;"></div> | ||
| - | We found that | + | We found that use of cellulose-treated epidermal cell sheet from onion is useful for quick evaluation of basic ability of ''E. coli'' expressing T3SS as protein injector. |
</div> | </div> | ||
{{Team:HokkaidoU_Japan/footer}} | {{Team:HokkaidoU_Japan/footer}} | ||
Latest revision as of 11:43, 15 December 2011
HokkaidoU Japan
iGEM 2011 Team of Hokkaido University
Contents |
- Abstract
- What`s T3SSDetailed information about T3SS and summary of our achievements on iGEM 2010
- Injection assay using onion cellsExperiments using plant cells are easier to perform than with mammalian ones
- Ready-to-inject backbone and Bsa I cloning siteReady-to-inject backbone and Bsa I cloning site enables easy fusion of T3S signal and protein
- GSK tag systemA neat injection assay using GSK tag, which can specifically detect successfully injected proteins
- Bsa I cloning site, RFC submissionDetailed documentation of costructing a BioBrick cloning site a BioBrick!
Injection assay using onion cells
SUMMARY
Last year, we have achieved injection of GFP to the RK13 cells (cultivated rabbit kidney cell) using E. coli expressing T3SS. However, using animal cells is inconvenient. For example, cultivation of the cells a must be started couple of days before infection. And deterioration of cultivation environment results in immediate death of cells. We thought that we might solve these problems by using onion cells. Onion has very widely available and its cells are tolerant of bacteria. Moreover, since onion forms detachable monolayer cell-sheets,they can be observe with ease. We repeatedly tried to establish injection protocol for onion cells and after some trials we were successful.
MATERIALS
- E. coli strain which can inject GFP into eukaryotic cells that we found last year.
- Epidermal cell sheet of onion
- Reagents
- MgM-MES pH7.2
- MgM-MES pH5.0
- 0.4 M Mannitol in MgM-MES pH5.0
- 1% cellulase 0.1% pectriase in 0.4M Mannitol solution (pH7.0)
- 20% L-arabinose solution
- Tetracyclin
- Chroramphenicol
PROCEDURE
Prepararion of bacterial suspension
- Isolate a single colony of E. coli and cultured cells in liquid LB (without antibiotics) for 2 hours.
- Add 500 ul of culture fluid to 2 ml of liquid LB. Add appropriate amount of antibiotics (Tetracycline and Chloramphenicol).
- Add 50ul of 20% L-arabinose solution.
- Culture cells overnight at 37C, shaking at 200rpm
- Spin down the cultured cells at 3000rpm, for 10 min at 35C. Remove supernatant.
- Add MgM-MES (pH5.0), Tetracycline, Chloramphenicol, L-arabinose to the bacterial pellet. Resuspend it using vortex mixer. Culture cells at 37C, shaking at 200rpm for 4 hours. This step is required because genes encoding salmonella T3SS are expressed in acidic environment.
- Spin down the culture fluid at 3000rpm, for 10 min at 25C. Remove supernatant.
- Resuspend the pellet with 2 ml of MgM-MES (pH5.0) containing 0.4 M Mannitol, then spin down the culture fluid at 3000rpm, for 10 min at 25C. Remove supernatant. Repeat this step 3 times. (This is done to remove the toxic substances that are produced by E. coli and adjust the osmotic pressure to that in onion cells)
- Resuspend the pellet with 2 ml of MgM-MES (pH5.0) containing 0.4 M Mannitol, Tetracycline, chloramphenicol and L-arabinose.
- Measure the light absorbance at 600 nm using a spectrophotometer, then adjust the concentration of cells to ΔOD600 = 0.06 by diluting with the same medium mentioned above.
- To infect onion cells with E. coliadd 500 ul of the culture fluid onto pre-treated onion cell-sheets. Leave at RT in petri dish (Prevent drying)
- Remove the bacterial culture fluid carefully by micropipette so that the cell-sheets are not to be torn. Observe the cells under a fluorescencent microscope.
Preparation of onion cells
Since we thought that the cell wall may prevent injection of protein, onion cell-sheets were pretreated to remove cell wall as follows.
- Cut an onion into four pieces.
- Take out a fresh sheets of epidermal cells (second or third layer from the surface of onion).
- Cut the layer into rectangular shape (about 1.5cm x 3cm)
- Tear off a cell sheet from the inner surface of the layer.
- Put the sheet onto a slide glass at back face up because the upper face deflects enzyme solution, and fix the edge of the sheet onto slide glass with 1% agarose gel.
- Fill a petri dish with 1% cellulase and 0.1% pectolyase in 0.4 M Mannitol solution (pH7.0), then submerge the cell sheet fixed on slide glass into the enzyme solution. Incubate for 8 min at RT.
- Prepare 3 petri dishes filled with MgM-MES pH5.0 containing 0.4 M Mannitol. After processing, wash cells stepwise soaking in these petri dishes to remove enzyme solution.
- Remove excess moisture, and transfer the sample to another petri dishs for infection.
Results
We carried out infection assay using following E. coli:
- T3SS signal(+)-GFP_T3SS signal(-)-RFP
- T3SS signal(-)-GFP
- T3SS only
Here are the results of infection assay figure 1 to 7. Figure 1 to 3 shows the results of infection assay using the E. coli which we have made last year. That E. coli has T3SS, GFP with secretion signal, and RFP.
As shown in following pictures GFP not fused to secretion signal is not injected to the onion cells.
We found that use of cellulose-treated epidermal cell sheet from onion is useful for quick evaluation of basic ability of E. coli expressing T3SS as protein injector.
 "
"