Team:Harvard/Technology/Chip Synthesis
From 2011.igem.org
Bioinformatics | Chip Synthesis | Plasmid Construction | Selection Strain Engineering | Selection Results
Chip Synthesis
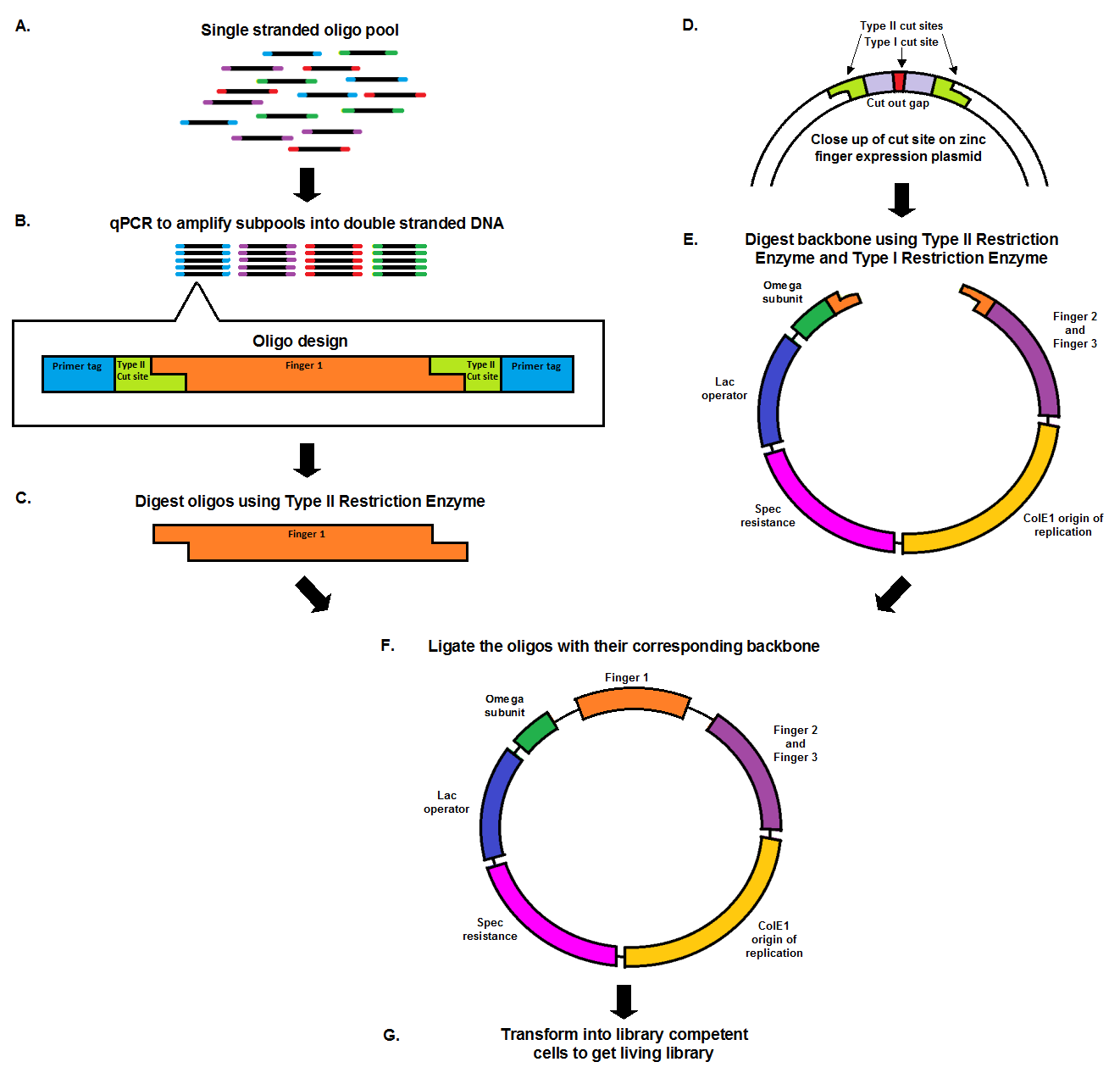
From Oligo Pool to Living Library
Chip synthesis results in a pool of single stranded oligos (A), designed with primer tags, which allow for the amplification of specific sub-pools, and Type II cut sites. qPCR, which amplifies the oligos at equal rates, results in the desired sub-pool (B). In order to prepare the oligo inserts and expression plasmid backbone for ligation, both must be cut with a Type II restriction enzyme (C, E). The cuts result in complementary sticky ends that anneal to each other during the ligation. The backbone, designed with a second cut site in the cut out gap (D), also undergoes a second digestion with a Type I restriction enzyme in order to minimize the number of undigested, or partially digested backbones. The oligo pool and backbone are then ligated (F), and transformed into library competent cells, resulting in a living library (G).
 "
"