Team:Harvard/Results/Zinc Finger Binders
From 2011.igem.org
(Difference between revisions)
(→Color Blindness (CB) Bottom Hits) |
|||
| Line 8: | Line 8: | ||
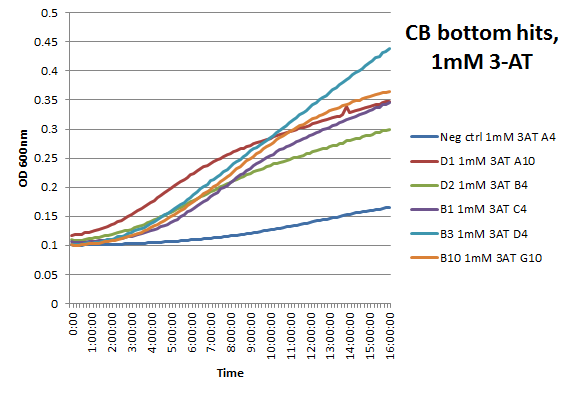
We assembled the Color Blindness Bottom library, transformed it into the proper selection strain, and plated on incomplete media with varying concentrations of 3-AT. We saw colonies form, with more on the less stringent plates (NM) and fewer colonies on the higher 3-AT concentrations. Several colonies were picked and tested under varying concentrations of 3-AT in plate reader overnight, and there was a clear difference in growth between the colonies and the negative control (CB bottom binding site with Zif268). After sequencing, we identified xx novel zinc fingers! | We assembled the Color Blindness Bottom library, transformed it into the proper selection strain, and plated on incomplete media with varying concentrations of 3-AT. We saw colonies form, with more on the less stringent plates (NM) and fewer colonies on the higher 3-AT concentrations. Several colonies were picked and tested under varying concentrations of 3-AT in plate reader overnight, and there was a clear difference in growth between the colonies and the negative control (CB bottom binding site with Zif268). After sequencing, we identified xx novel zinc fingers! | ||
| - | [[File: HARVcbbot_1mM.png]] | + | [[File: HARVcbbot_1mM.png|frameless]] |
</div> | </div> | ||
Revision as of 20:42, 25 September 2011
Overview | MAGE | Lambda Red| Chip-Based Library | One-Hybrid Selection | Zinc Finger Binders | Biobricks
Color Blindness (CB) Bottom Hits
We assembled the Color Blindness Bottom library, transformed it into the proper selection strain, and plated on incomplete media with varying concentrations of 3-AT. We saw colonies form, with more on the less stringent plates (NM) and fewer colonies on the higher 3-AT concentrations. Several colonies were picked and tested under varying concentrations of 3-AT in plate reader overnight, and there was a clear difference in growth between the colonies and the negative control (CB bottom binding site with Zif268). After sequencing, we identified xx novel zinc fingers!
 "
"