Team:Harvard/Project
From 2011.igem.org
| Line 18: | Line 18: | ||
As the structure and binding interactions of zinc fingers are not yet understood, our project utilizes bioinformatics and computational analysis of the limited existing data to make “educated guesses” of what amino acid sequences will bind to our desired target sequence. | As the structure and binding interactions of zinc fingers are not yet understood, our project utilizes bioinformatics and computational analysis of the limited existing data to make “educated guesses” of what amino acid sequences will bind to our desired target sequence. | ||
| - | We chose 6 target sequences in the human genome that no currently-existing zinc finger is able to bind to: genes that cause colorblindness, some types of cancer, and high cholesterol. These targets were chosen after conducting an extensive literature search, with the most useful information coming from a previous zinc finger study called CODA<sup> | + | We chose 6 target sequences in the human genome that no currently-existing zinc finger is able to bind to: genes that cause colorblindness, some types of cancer, and high cholesterol. These targets were chosen after conducting an extensive literature search, with the most useful information coming from a previous zinc finger study called CODA<sup>[[#References|[3]]]</sup>. |
We emailed the OPEN zinc finger consortium and Dr. Anton Persikov to acquire their respective databases of zinc fingers: Persikov had compiled the results of the past 10 years of zinc finger research. We then decided how to analyze this data and choose targets for new zinc fingers (details below). Then we programmed our ideas into a Python program, and generated 55,000 potential zinc fingers. | We emailed the OPEN zinc finger consortium and Dr. Anton Persikov to acquire their respective databases of zinc fingers: Persikov had compiled the results of the past 10 years of zinc finger research. We then decided how to analyze this data and choose targets for new zinc fingers (details below). Then we programmed our ideas into a Python program, and generated 55,000 potential zinc fingers. | ||
Revision as of 00:12, 19 October 2011
Overview | Design | Synthesize | Test | Zinc Finger Background | Protocols
Contents |
Our Project
Zinc fingers are specialized proteins that bind to DNA. Due to their ability to target highly specific DNA sequences, zinc fingers offer great potential for gene therapy and personalized medicine: recently, they were shown to be effective in conferring HIV resistance and treating hemophilia in mice. In the past, however, designing new zinc fingers - a necessity for individualized gene therapy - has been prohibitively expensive and time consuming.
For our project, we created and tested thousands of zinc fingers at a cost feasible for most labs. To do so, we harnessed two novel synthetic biology technologies: chip-based synthesis1, which allows for thousands (even millions) of DNA strands to be synthesized concurrently, and multiplex automated genome engineering (MAGE)2, which makes possible direct edits of the genome of organisms, rather than using small, cumbersome plasmids.
To do this, our project has three main steps:
1. Design
Use a bioinformatics approach to predict 55,000 zinc finger sequences.
As the structure and binding interactions of zinc fingers are not yet understood, our project utilizes bioinformatics and computational analysis of the limited existing data to make “educated guesses” of what amino acid sequences will bind to our desired target sequence.
We chose 6 target sequences in the human genome that no currently-existing zinc finger is able to bind to: genes that cause colorblindness, some types of cancer, and high cholesterol. These targets were chosen after conducting an extensive literature search, with the most useful information coming from a previous zinc finger study called CODA[3].
We emailed the OPEN zinc finger consortium and Dr. Anton Persikov to acquire their respective databases of zinc fingers: Persikov had compiled the results of the past 10 years of zinc finger research. We then decided how to analyze this data and choose targets for new zinc fingers (details below). Then we programmed our ideas into a Python program, and generated 55,000 potential zinc fingers.
2. Synthesize
Use chip-based DNA synthesis to make 55,000 sequences simultaneously, then insert the oligos into E.coli.
3. Test
Use a metabolic selection system to test which zinc finger sequences successfully bind DNA.
Thus, our zinc fingers and their clinical applications are a new technology, and while we hope that our new zinc fingers work, the more important goal is maximizing efficiency and decreasing cost while utilizing new technology: we anticipate that future iGEM teams will find great use for chip-based synthesis and MAGE.
See here for our abstract and detailed project description.
Technological Applications
The novel methods we employed in our project have the potential to revolutionize synthetic biology practices, and the way that future iGEM competitions are conducted. To learn more about the technological applications of our project, please see our Technology page.
Zinc Finger Background
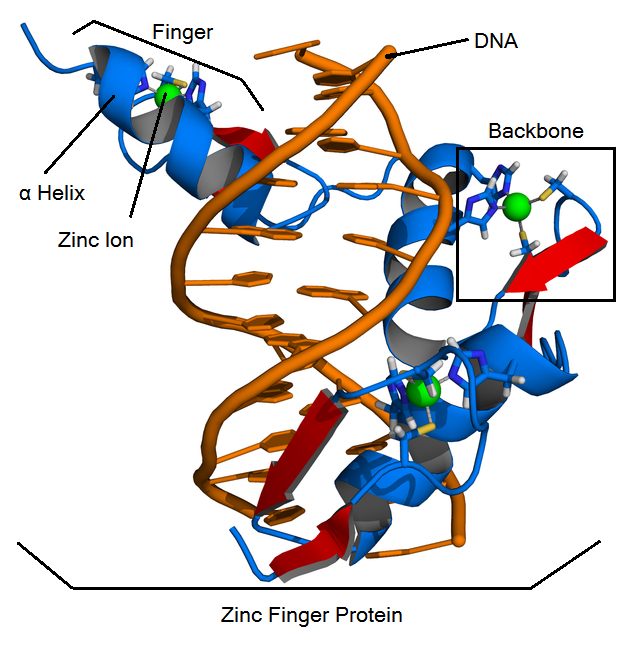
What are Zinc Finger Proteins (ZFPs)?
Function
ZFPs are found commonly in nature as a class of special transcription factors that bind to DNA, thus regulating gene expression. Zinc finger function was first studied using zinc finger protein Zif268.
Structure
ZFPs consist of smaller subunits called "fingers" which each contain a zinc finger binding helix that binds to unique DNA sequences. These fingers are linear and linked together by the "zinc finger backbone", a series of approximately 21 amino acids.
- Cis2His2 ZFPs have three main structural components:
- Zinc finger binding helix
- Linker region
- Zinc ion that is coordinated by two cysteine residues and two histidine residues.
Helpful Zinc Finger Links
[http://en.wikipedia.org/wiki/Zinc_finger Zinc Fingers on Wikipedia]
- A more detailed introduction to zinc fingers.
[http://compbio.cs.princeton.edu/zf/ Predicting DNA Recognition by C2H2 Zinc Finger Proteins]
- A program useful for predicting how well a given amino acid sequence will bind to a given DNA sequence
[http://www.zincfingers.org/default2.htm The Zinc Finger Consortium]
- Information & helpful resources for zinc fingers
[http://www.jounglab.org/ Joung Lab]
- Information about Dr. Joung's extensive work with zinc fingers
Clinical Applications of Zinc Fingers
Colorblindness (Red Opsin)
- Goal: Produce functional red opsin photoreceptor proteins in the eye
- Method: Insertion of functional red opsin gene (OPN1LW [http://genome.ucsc.edu/cgi-bin/hgGene?hgg_geneuc004fjz.3&hgg_protP04000&hgg_chromchrX&hgg_start153409724&hgg_end153424505&hgg_typeknownGene&dbhg19&hgsid206379197 1]) upstream of normal locus in patient lacking the gene
Inherited High Cholesterol (Familial Hypercholesterolemia)
- Goal: Produce functional LDLR protein to remove LDL cholesterol from the blood
- Method: Insertion of functional LDLR gene upstream of nonfunctional allele
Cancer (Myc Oncogene)
- Goal: Knock out the oncogenic protein product and stop cancerous proliferation
- Method: Targeted disruption (deletion) in mutated oncogene
References
1. Harris H. Wang, Farren J. Isaacs, Peter A. Carr, Zachary Z. Sun, George Xu, Craig R. Forest, George M. Church. Programming cells by multiplex genome engineering and accelerated evolution. (2009). Nature, 460(7257):894-8. [http://www.nature.com/nature/journal/v460/n7257/full/nature08187.html]
2. Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, Emig CJ, Bang D, Hwang SJ, Jewett MC, Jacobson JM, Church GM. (2011). Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science, 333(6040):348-53. [http://www.sciencemag.org/content/333/6040/348.short]
3. Segal, David J. Zinc-finger nucleases transition to the CoDA (2011). Nature Methods 8, 53–55. [http://www.nature.com/nmeth/journal/v8/n1/full/nmeth0111-53.html]
 "
"