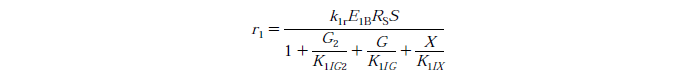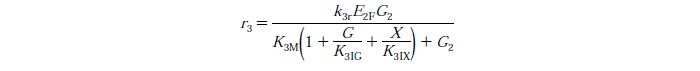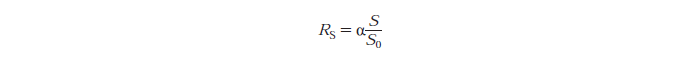Team:Edinburgh/Cellulases (MATLAB model)
From 2011.igem.org
Cellulases (MATLAB model)
The most important part of the biorefinery is the reactor where cellulose is converted to glucose. But accurately predicting how much is converted, using synergy between enzymes is difficult without literature to provide the ordinary differential equations (ODE's) and the kinetic parameters. Therefore this model only looks at the free floating enzyme approach (non-synergy). It is deterministic i.e non random and is set by a series of initial conditions.
Contents |
Assumptions
The mathematical model is based on the ODE's and kinetic parameters outlined in [http://onlinelibrary.wiley.com/doi/10.1021/bp034316x/full Kadam et al, 2004]. The following are its assumptions and basis:
- Rate equations assume enzyme adsorption follows the Langmuir isotherm model
- Glucose and cellobiose which are the products of cellulose hydrolysis, were assumed to, 'competitively inhibit enzyme hyrolysis' [http://onlinelibrary.wiley.com/doi/10.1021/bp034316x/full Kadam et al, 2004]
- Assume all reactions follow the same temperature dependency Arrhenius relationship. However it should be different for every enzyme component, 'because of their varying degrees of thermostability, with beta-glucocidase being the most thermostable. Hence the assumption is a simplification of reality' [http://onlinelibrary.wiley.com/doi/10.1021/bp034316x/full Kadam et al, 2004]
- Conversion of cellobiose to glucose follows the Michaelis-Menten enzyme kinetic model
Equations
Rate Equations
Constants
knr - reaction rate constant for reaction n
EnB is the bound concentration for exo and endo-beta-1,4-glucanase for reaction n
Rs - substrate reactivity parameter
S - substrate reactivity at a given time (g/kg)
G2 - concentration of cellobiose
G - concentration of Glucose
X - xylose concentration
KnIG2 - inhibition constant for cellobiose at reaction n
K1IG - inhibition constant for Glucose at reaction n
K1IX - xylose inhibition constant for reaction n
Note: Assuming no xylose inhibition therefore X=0
Langmuir Isotherm
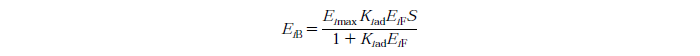
Constants
exo and endo-beta-1,4-glucanase, i=1
beta-glucosidase, i=2
Eimax - Maximum mass of exo and endo-beta-1,4-glucanase (i=1) or beta-glucosidase (i=2) that can be absorbed onto a unit of mass substrate
Kiad - Dissociation constant for enzyme i
EiF - Free enzyme concentration for enzyme i
S - Substrate reactivity at a given time (g/kg)
Mass Balances
Arrhenius Equation
Constants
Kir - Reaction rate constant of reaction i
Eai - Activation energy of reaction i
R - Universal gas constant
Construction of Model
The model was constructed using the numerical programme [http://www.mathworks.co.uk/products/matlab/index.html MATLAB]. A script file was generated which holds the variable dictionary, constants, temperature dependency equations, ODE45 and plot command.
A separate function file to the script is created as script files can only operate on the variables that are coded into their m-files. Rs, the substrate reactivity parameter changes at every iteration because it is dependent on the S the substrate concentration at a given time, the substrate being cellulose. Therefore S at the first iteration is So, the initial substrate concentration. At the second iteration S is the previous value calculated by the ODE, and so on. After each step the new value of S, is fed into the function file and is used for calculating the reaction rate constant for cellobiose and glucose etc.
ODE45 calls on the function file to calculate the rate equations and then substitute them into the respective mass balance. A numerical integration is performed and the results can be seen below. ODE 45 is used as it is more accurate than other solvers [http://www.mathworks.co.uk/help/techdoc/ref/ode23.html (Mathworks)]. It is based on an explicit Runge-Kutta formula and is a one step solver.
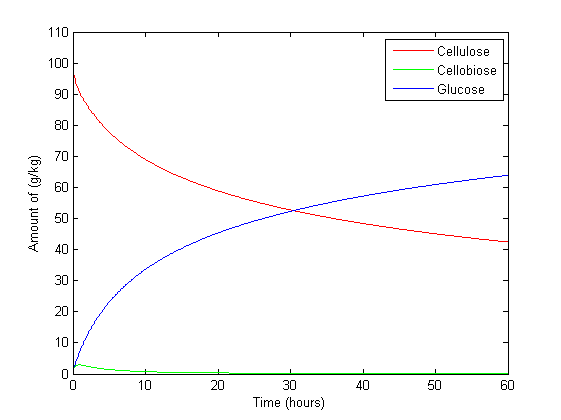
Results
The above graph is set with beta-glucosidase at its maximum concentration of 1g/kg and exo/endo glucanase at its lowest to show what effects different enzyme concentrations has cellulose, glucose and cellobiose levels. The result is consistent with what is expected. β
References
- Kadam KL, Rydholm EC, McMillan JD (2004) [http://onlinelibrary.wiley.com/doi/10.1021/bp034316x/full Development and Validation of a Kinetic Model for Enzymatic Saccharification of Lignocellulosic Biomass]. Biotechnology Progress 20(3): 698–705 (doi: 10.1021/bp034316x).
 "
"