Team:Edinburgh/Cellulases (Kappa model)
From 2011.igem.org
| Line 66: | Line 66: | ||
Since Kappa models can be easily extended (by adding new agents, or reaction rules), we tried to make a synergistic model based on the non-synergistic model. | Since Kappa models can be easily extended (by adding new agents, or reaction rules), we tried to make a synergistic model based on the non-synergistic model. | ||
| - | [[Edinburgh_Inp.png|center|thumb|700px|A diagram of the spatial model.]] | + | [[File:Edinburgh_Inp.png|center|thumb|700px|A diagram of the spatial model.]] |
TODO: PICTURE HERE OF INP_EXO_ENDO_BGLU | TODO: PICTURE HERE OF INP_EXO_ENDO_BGLU | ||
Revision as of 14:07, 29 August 2011
Contents |
Kappa
Kappa is a stochastic, agent-based language for simulating interactions in (mostly) biological systems. It has been used by several iGEM teams before, most notably by Team Edinburgh 2010, who won a Best Model Prize for it.
Advantages of Kappa:
- easy to learn - you can summarise the whole language on one page
- no need for knowing mathematical formulae (ODEs) - the sophisticated code does that for you
- the models are relatively easy and quick to develop
- there exist some graphical tools to aid creation of models - e.g. RuleBase
Kappa represents the world in terms of agents. Agents in Kappa can have states, and can be linked to one another by bonds. Reactions are represented as creation/deletion of agents/bonds.
Structure of the model
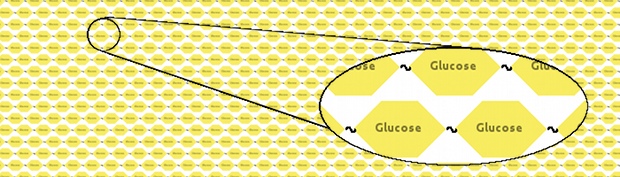
We thought that a neat model would represent cellulose as a long fibre of glucoses linked together:
However, manually inputting that would be a considerable effort. Instead, we made a Kappa system which would build cellulose for us, and then, using Kappa syntax, took a snapshot of the model. Two variants were created, one with long (10,000 glucoses) fibres of cellulose and another one with short (1,000 glucoses) fibres.
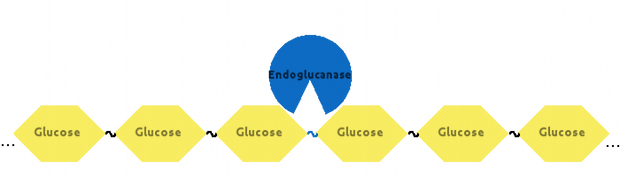
Then, Endoglucanase would cut Cellulose somewhere in the middle:
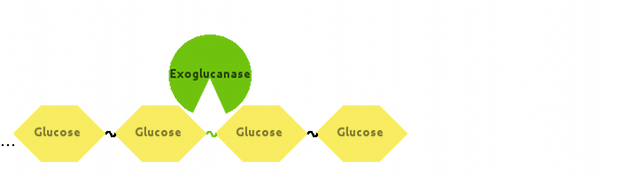
Also, Exoglucanase would cut Cellulose at end-points:
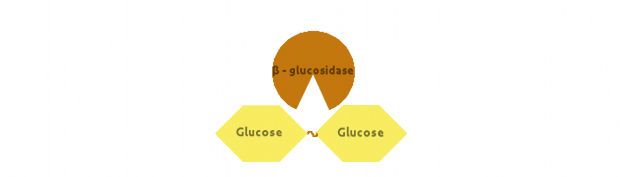
Finally, β-glucosidase would cut dimers (i.e. Cellobiose):
The exact reaction rules are available as a file here.
Non-synergistic model
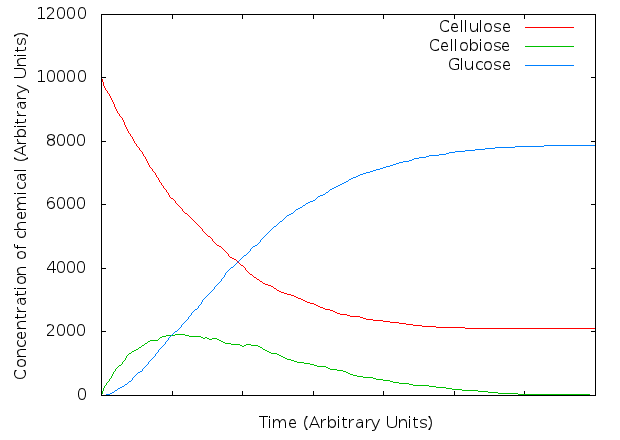
We developed our first, non-synergistic model in Kappa, based on the system as seen above. A preliminary model showed similar graphs to those in Kadam, Rydholm & McMillan (2004). However, after closer examination, the model made us think of one problem - glucose trimers.
Glucose trimers
The first slightly unanticipated result of our simulations was that, depending on how the model is set up, trimers of glucose can appear. If none of the (simulated) enzymes can degrade this, it will be one of the terminal products.
The following graph shows concentrations of glucose, cellobiose, and "cellulose" (which is defined as any glucose chain of length greater than 2) in a simple simulation where all three cellulase types are present but none is capable of degrading the trimer... at the end, all of the so called "cellulose" is composed of these trimers.
This immediately led us to the question of whether any of our three enzymes (Cellulomonas fimi endoglucanase, exoglucanase, and β-glucosidase) are in fact capable of degrading the trimer...
Improved model
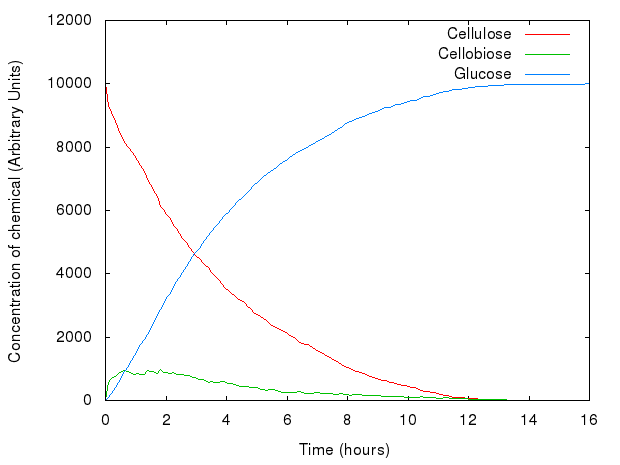
For the rest of our models, we assumed that some of our enzymes CAN degrade such trimers.
Parameter fitting
We carried out a series of experiments in order to fit parameters to our model. The aim was to generate graphs similar to those in Kadam, Rydholm & McMillan (2004) (since back then we did not have any data available from the wet lab). We manipulated the kinetic rates of reactions and relative concentrations of enzymes.
After an overnight series of experiments, we came up with a range of values for parameters of our model.
Synergy model
Since Kappa models can be easily extended (by adding new agents, or reaction rules), we tried to make a synergistic model based on the non-synergistic model.
TODO: PICTURE HERE OF INP_EXO_ENDO_BGLU
Why it failed?
PICTURE HERE
Spatial Kappa
Spatial Kappa is an extension to Kappa made by our last year's team (Team Edinburgh 2010). It seems like the extra functionality of compartments added by Spatial Kappa would help develop a model of synergistic action.
TODO: Synergy - enzymes don't move. Non-Synergy - enzymes move around.
References
- Kadam, KL, Rydholm, EC, McMillan JD (2004) Development and Validation of a Kinetic Model for Enzymatic Saccharification of Lignocellulosic Biomass. Biotechnology Progress Vol. 20, Issue 3: 698–705, (doi: 10.1021/bp034316x).
 "
"