Team:ETH Zurich/Biology/Validation
From 2011.igem.org
(→AraC - the arabinose sensor) |
(→Experimental results) |
||
| Line 4: | Line 4: | ||
= Experimental results = | = Experimental results = | ||
'''For the 3 sub part of SmoColi we designed additional testsystems to test the functionality of the single parts. Alike all new or improved biobricks were tested and characterized before they were implemented in the sub parts.''' For the sensors we tested the repressors or activators in presents of the corresponding small molecule. For the P<sub>u</sub> promoter show a dose response for xylene over air, the same for the arabinose, by knocking out the transporter the ON-/OFF character of AraC could be changed to a nice dose response. The funghi AlcR sensor is a element which can not be used in ''E.coli''. | '''For the 3 sub part of SmoColi we designed additional testsystems to test the functionality of the single parts. Alike all new or improved biobricks were tested and characterized before they were implemented in the sub parts.''' For the sensors we tested the repressors or activators in presents of the corresponding small molecule. For the P<sub>u</sub> promoter show a dose response for xylene over air, the same for the arabinose, by knocking out the transporter the ON-/OFF character of AraC could be changed to a nice dose response. The funghi AlcR sensor is a element which can not be used in ''E.coli''. | ||
| + | [[File:File:ETHZ_green_band.jpg|300px|left|thumb|'''Figure 1: It's not what you think it is! *''']] | ||
{{:Team:ETH Zurich/Templates/SectionEnd}} | {{:Team:ETH Zurich/Templates/SectionEnd}} | ||
Revision as of 20:09, 28 October 2011

Contents |
Experimental results
For the 3 sub part of SmoColi we designed additional testsystems to test the functionality of the single parts. Alike all new or improved biobricks were tested and characterized before they were implemented in the sub parts. For the sensors we tested the repressors or activators in presents of the corresponding small molecule. For the Pu promoter show a dose response for xylene over air, the same for the arabinose, by knocking out the transporter the ON-/OFF character of AraC could be changed to a nice dose response. The funghi AlcR sensor is a element which can not be used in E.coli.
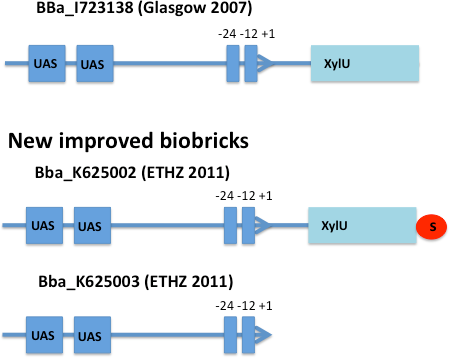
XylR - the xylene sensor
E. coli strain JM101 was transformed with two plasmids containing the transcriptional regulator XylR, the degradation cassette xylMABN and a GFP reporter coupled to BBa_K625002 respectively BBa_K625003. BBa_K625002 is a improved version of the Pu promoter BBa_I723020 were we inserted a stop codon to prevent fusion proteins. BBa_K625003 is a minimal version of the Pu promoter were the whole XylU part is deleted. For the first two of those we used the plasmid pCK04AxylR according to [4]. The reporter plasmid we constructed ourselves by using BBa_K625005 as a backbone.
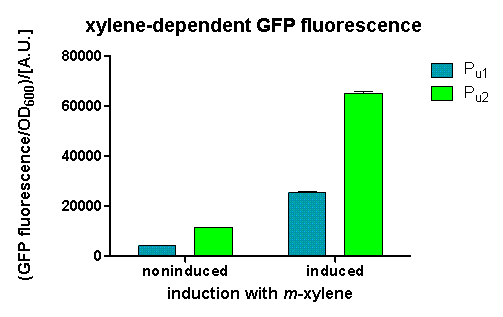
We inoculated 50 mL of LB medium with an overnight culture of the co-transformed strains and set the OD600 to 0.1. Upon reaching the exponential growth phase, the cultures were induced with m-xylene. Therefore we put a sterile test tube with m-xylene or a mixture out of m-xylene and paraffin oil into the flask and sealed it with parafilm in order to get an air-induced response.
OD600 and GFP fluorescence was measured after different time periods in a 96-well plate. Afterwards the data was normalized in order to get more consistent results.
Comparison of the two promoter versions
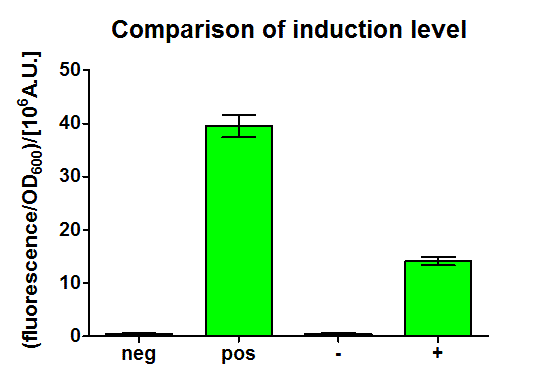
After 3 h we could observe a clear increase in GFP fluorescence when m-xylene was added in a test tube. Thus GFP production can be induced by m-xylene present from the air. The results of the experiments can be seen in Figure 8. By comparing [http://partsregistry.org/Part:BBa_K625002 BBa_K625002] and [http://partsregistry.org/Part:BBa_K625003 BBa_K625003], we can see that the leaky expression of GFP is higher in the shortened promoter [http://partsregistry.org/Part:BBa_K625003 BBa_K625003] compared to the longer one. There are several possible reasons for this observation. Most likely it is due to the drop of the natural part downstream of the promoter, which is usually also included in Pu.
Nevertheless, we could also see a higher level of induction in the setup with [http://partsregistry.org/Part:BBa_K625003 BBa_K625003] (around 7-fold) compared to the [http://partsregistry.org/Part:BBa_K625002 BBa_K625002] setup (around 5-fold). Thus both of the provided promoters can be used for transcriptional regulation with XylR.
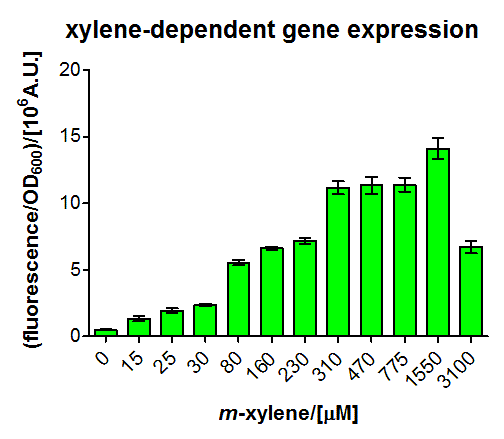
Results of BBa_K625003 characterization
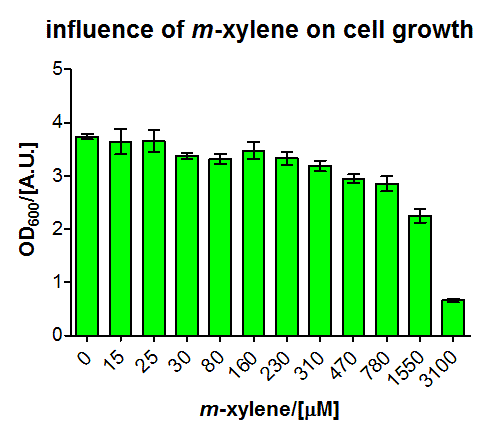
For the short version of the PU promoter we performed a more detail characterization. We measured GFP induction with different xylene concentrations by adding different mixtures of xylene and paraffin oil into the test tube. (Calculations for xylene concentration in the medium) In previous experiment done in LB medium we saw that the PU promoter is leaky, so we did the experiment with M9 with 0.5 % glucose. We measured after fluorescence and OD600 after 8 h. Xylene has a toxic effect on E.coli 3.1 mM are toxic for the cells, already 1.55 mM cell growth is inhibit. The maximal GFP production is between 3 and 7 mM.
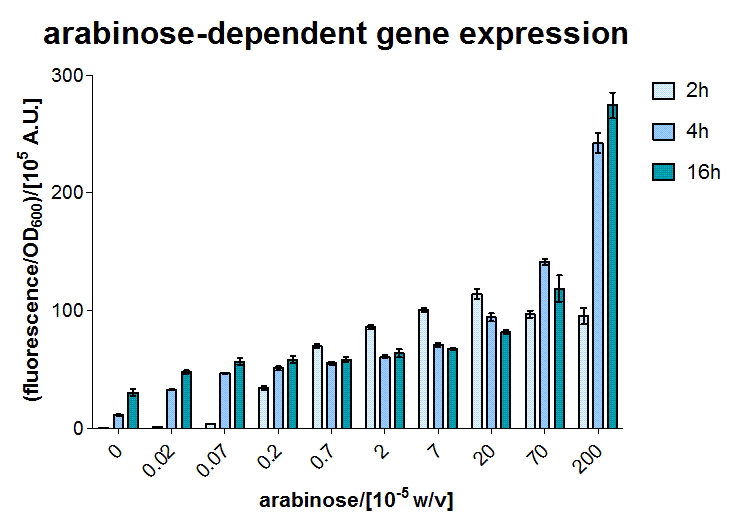
AraC - the arabinose sensor
A E.coli BW27783 stain was transformed with a plasmid containing GFP reporter coupled to the PBAD promoter, which can be activated by arabinose bound AraC. AraC is included on the genome. In the exponential phase our BW27783 E.coli were induced with several arabinose concentrations.
AlcR- the acetaldehyde sensor
As described in the Smoke detectors section we designed two artificial AlcR-dependent promoters which get repressed upon binding of AlcR to acetaldehyde. For testing of our design we created a system with a sfGFP output in order to characterize the gene expression with different levels of AlcR production and acetaldehyde induction.
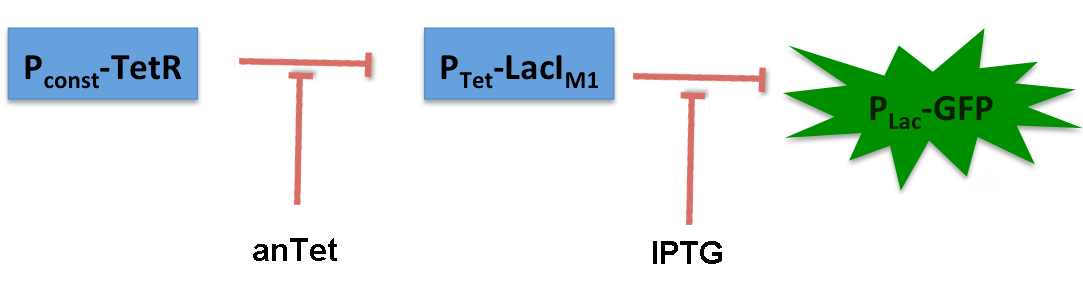
The system works in the following way (see Figure 9):
A) In normal medium conditions the TetR transcription factor is produced and binds to its cognate promoter, thus inhibiting the expression of alcR. In this case no AlcR is present and GFP is produced.
B) Upon addition of increasing amounts of anhydrotetracycline, TetR gets released from the promoter and AlcR is produced. The amount of AlcR should increase with increasing amounts of anhydrotetracycline.
C) By addition of acetaldehyde AlcR binds to the alc-promoter and the GFP response decreases.
Experiments and results for funghi AlcR
In a first experiment, the alc-promoters were tested by using a non-codon optimized natural variant of alcR. Because of this we tested the compatibility of the codon usage in the variant from Aspergillus nidulans with the one from E. coli. We received a codon adaptation index (CAI) of 0.7 [2]. Nevertheless, five rare arginine codons in E.coli were present within the first 20 amino acids of the gene. After a first experiment without any significant results (data not shown), the first three rare arginine codons of alcR were exchanged by PCR and a fully codon-optimized variant was ordered. All of the following experiments were performed with alcR codon-optimized within the first 20 amino acids.
The AlcR test was established as a two plasmid system containing ampicillin and kanamycin resistances and pMB1 respectively pSC101 origins of replication. These plasmids were co-transformed into E. coli JM101 strain. M9 medium was inoculated with an overnight culture of the strain. The bacteria were induced with different amounts of acetaldehyde (0, 10, 1000 µM) and anhydrotetracycline (0, 1, 100 ng/mL) during the exponential growth phase. In order to avoid evaporation of acetaldehyde, all equipment was cooled down to -20 degrees prior to preparation of the stock solutions. All solutions were afterwards prepared in tubes at 4°C. Finally, the cell suspension was added to the solutions and the tubes were tightly closed to prevent loss of acetaldehyde. Fluorescence and OD measurements were performed in a 96-well plate several hours after induction.
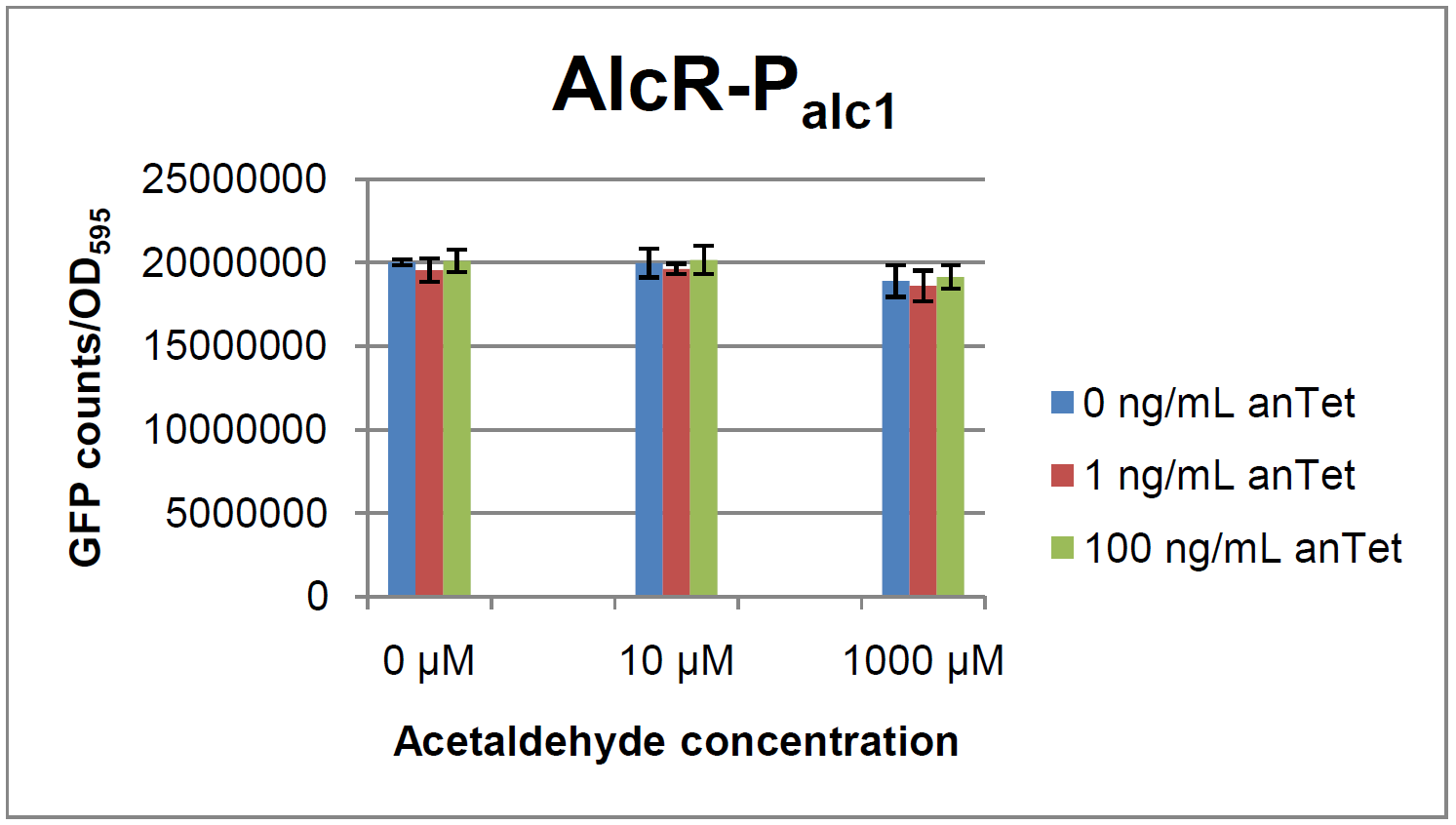
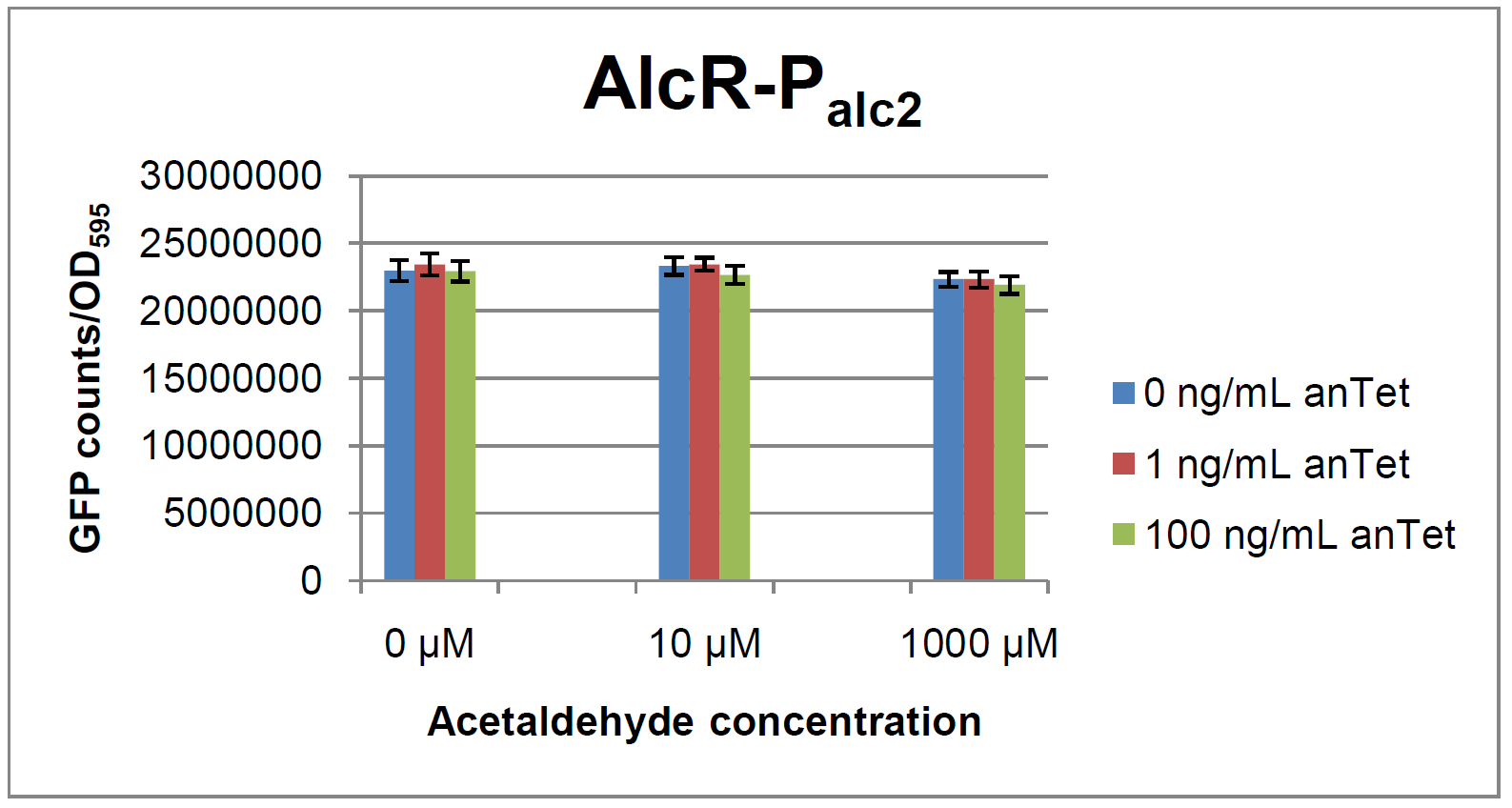
As visible on the two images above, no significant change in GFP fluorescence could be observed neither between different acetaldehyde concentrations nor between different AlcR induction levels. Because of these negative results and the non codon-optimized alcR, we performed expression tests for the protein in E. coli strain JM101. In these experiments, gene expression was induced with different amounts of anhydrotetracycline during the exponential growth phase of the bacteria. After cultivation, cells were harvested and lysed either by using a Retsch mill or by lysozyme. After resuspension of the cell extract we performed both SDS-PAGE gels and western blots to check for alcR expression. Buffer was added to the cell debris and heated to 95°C for 10 minutes in order to resolubilize potential inclusion bodies.
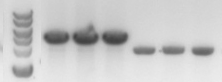
AlcR has a molecular mass of about 96 kDa and thus its band was suspected to be on the same level as the red box marked in the SDS-PAGE gel (see Figure 4) [3]. Despite increasing amounts of anhydrotetracycline (wells 1 and 3: 0 ng/mL, 2 and 4: 1 ng/mL, 3 and 6: 100 ng/mL), no change could be observed in this area of the gel for any of the probes. A reason for this could be that alcR is not expressed at all.
To check again whether alcR is expressed at all we performed a western blot. Therefore, a 6x His-tag was added to the C-terminal end of AlcR and we used an anti-His antibody kit in order to detect it. A probe with another concentrated 6x His-tagged protein was added as positive control so that we could exclude systematic errors during the blotting procedure (see Figure 5).
Because we could not show any expression of alcR at all we suppose that there is no AlcR produced in the cells, probably due to the usage of rare codons in the gene. Due to a long delay in the synthesis of the codon-optimized part, we were not able to test our system with this variant until now.
Bandpass-Characterization of lacIM1
LacIM1 ([http://partsregistry.org/Part:BBa_K625000 Bba_K625000]) is a LacI variant with the same protein sequence as the wild-type LacI, but with a completely codon-modified DNA sequence. [5] This enables to use both sequences on two different plasmids (with two different promoters) without recombination happening between them, which is necessary for in our system to establish the bandpass filter. By entering this BioBrick into the registry, we wanted to check its functionality. Thus we engineered a test system for our BioBrick by adding a lac-dependent GFP reporter. A Tet promoter (PTet) was put in front of lacIM1 and TetR constitutively produced, thus preventing lacIM1 from expression.
In the non-induced state of the system, lacIM1 expression is repressed and the cells produce GFP. If anhydrotetracycline is added, TetR is released from the Tet promoter (Ptet) and LacI is produced. LacI inhibits gfp transcription and the fluorescence decreases. Addition of IPTG although leads to the opposite case: LacI gets inhibited and the fluorescence increases.
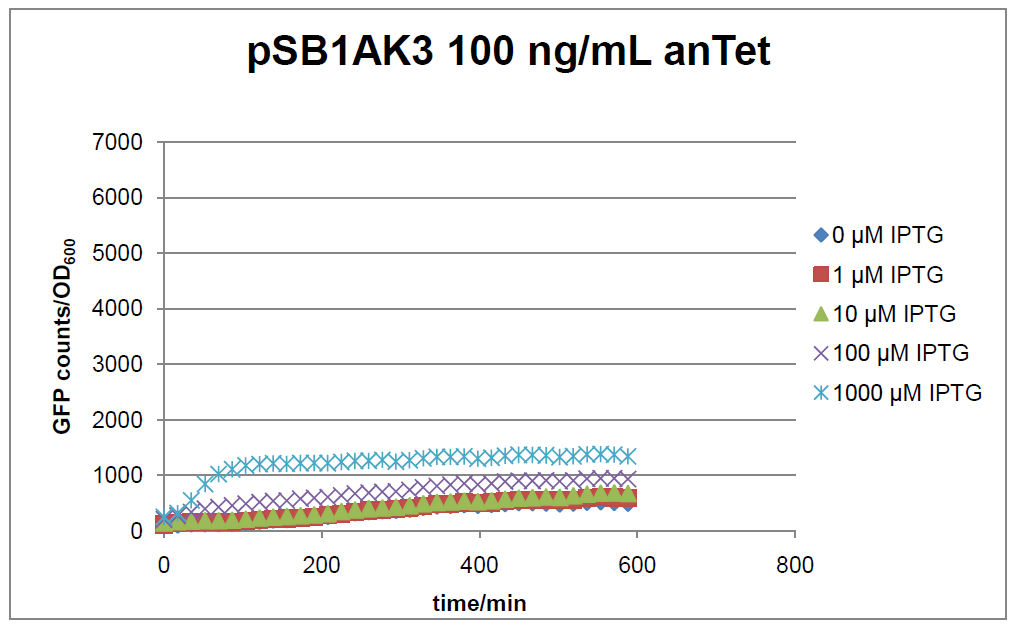
The test system for Bba_K625000 was established as a single plasmid system. In the experiments we used the two different plasmids pSB1AK3 and pSB3C5 as backbones of the designed network. Experiments were performed in E.coli strain JM101, which was grown in M9 medium until exponential growth and then induced with increasing amounts of anhydrotetracyline (0 ng/mL, 1 ng/mL, 100 ng/mL) and IPTG (0 µM, 1 µM, 10 µM, 100 µM, 1000 µM). The cells were incubated in a 96-well plate at 37°C and every 15 minutes OD600 and GFP fluorescence was measured.
Results
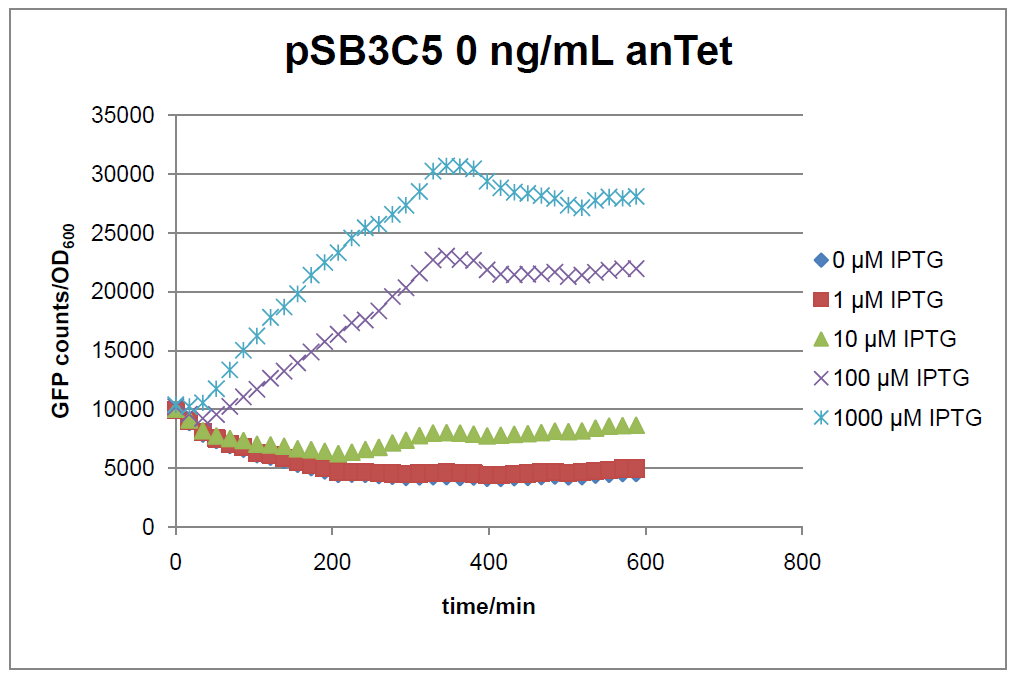
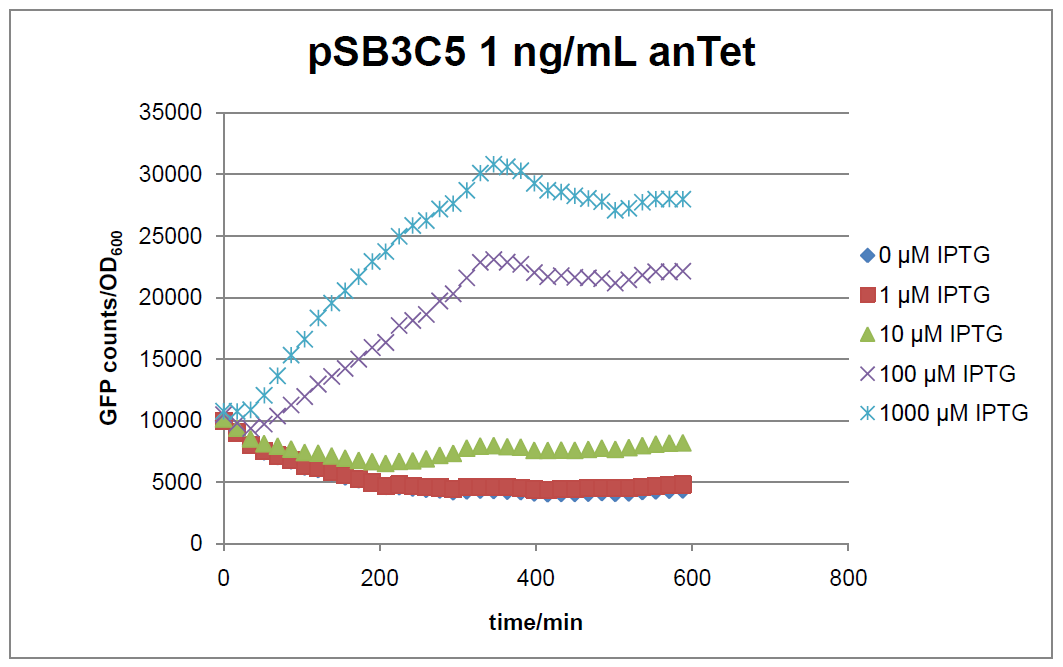
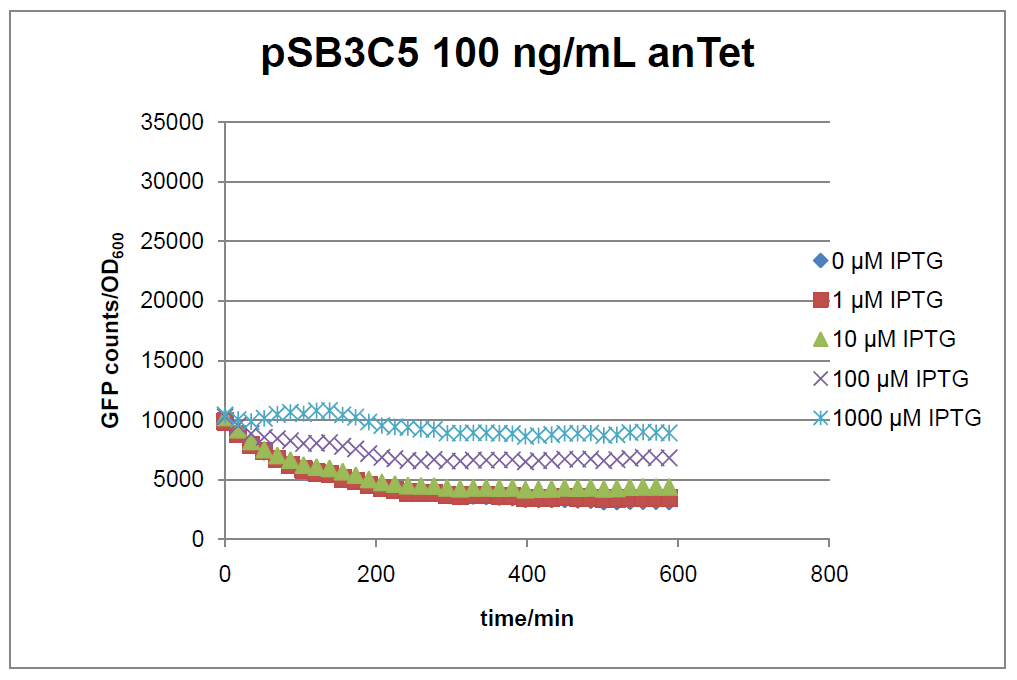
Figure 15: time course of fluorescence depending on different expression levels of the transcriptional regulator LacIM1. pSB3C5 was used as plasmid backbone for the test system.
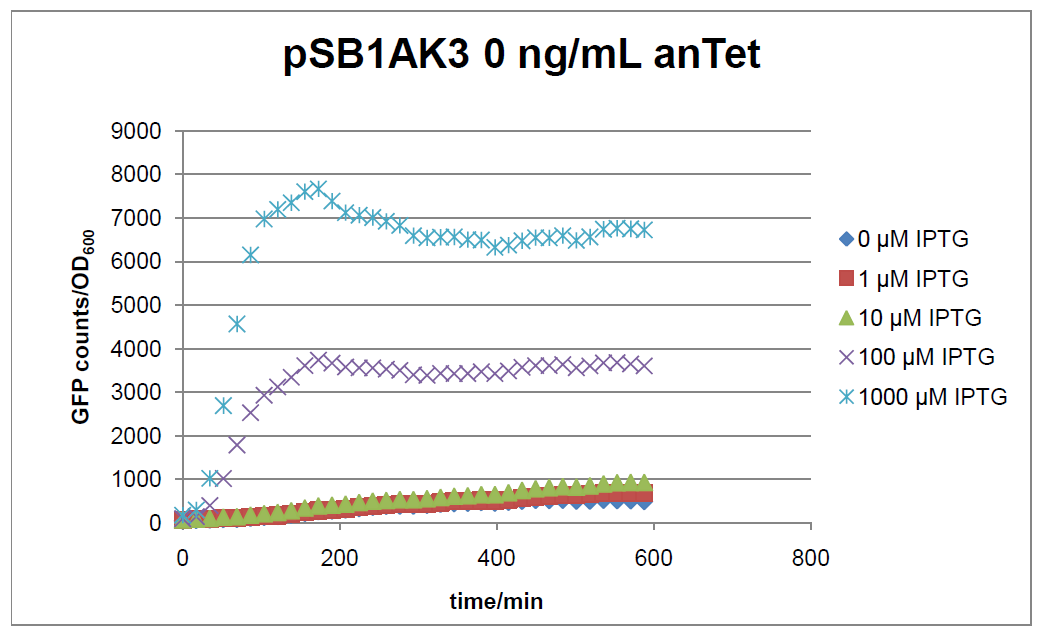
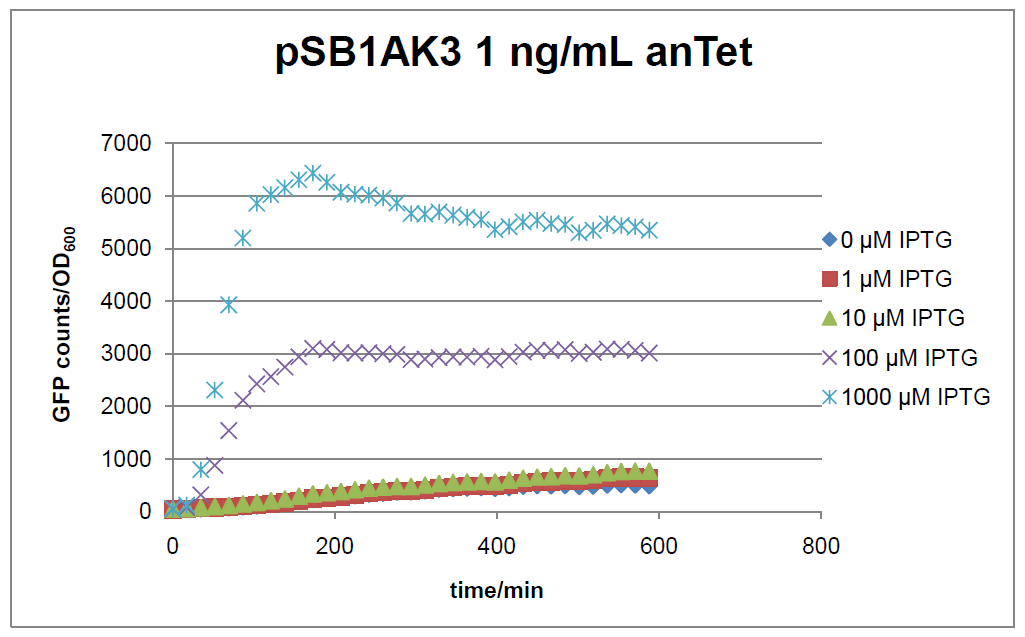
Figure 16: time course of fluorescence depending on different expression levels of the transcriptional regulator LacIM1. pSB1AK3 was used as plasmid backbone for the test system.
In general the test system for characterization of our BioBrick parts BBa_K625000 respectively BBa_K625001 worked as expected. In both system we can see that the fluorescence decreases with increasing amounts of anhydrotetracycline. This indicates that LacIM1 is induced and thus represses the expression of gfp. By addition of IPTG LacIM1 gets inhibited and therefore the GFP response increses. This tendency can be observed both in figure 15 and figure 16.
At a concentration of 100 ng/mL anhydrotetracycline, only a slight increase in fluorescence can be observed for increasing IPTG concentrations. We are supposing that the expression of lacIM1 is fully induced at this level and thus the IPTG amount present in the system is not sufficient anymore to inhibit all of the LacIM1.
In the case where no anhydrotetracycline is present in the medium, there can still be observed a change in fluorescence depending on the level of IPTG. The same pattern of induction can be observed here as in the weakly induced systems. The reason for this is likely due to the leaky expression of the lacIM1 gene, resulting in a small amount of LacIM1 always present in the cells and thus inhibiting the GFP production.
After about 300 min for pSB3C5 and about 200 min for pSB1AK3, the cells enter stationary phase and the flourescence intensity and OD600 do not increase anymore.
Alarm
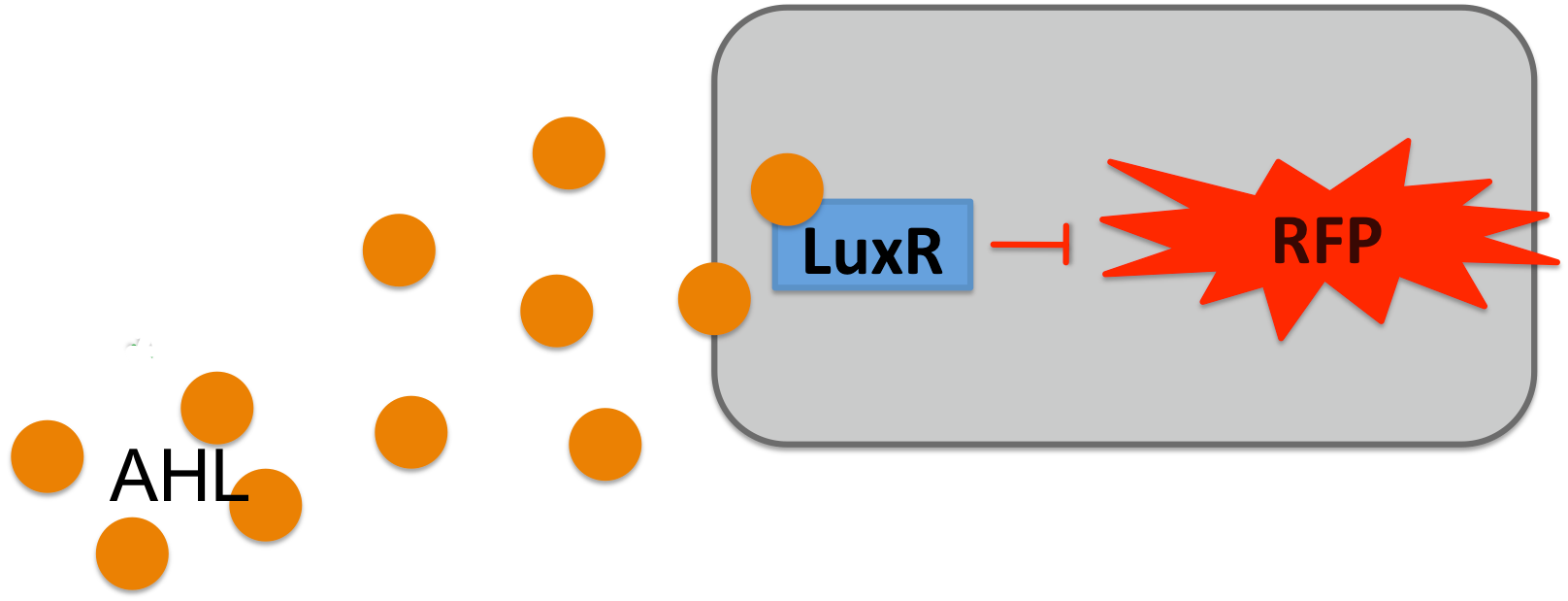
To test the alarm sytsem we induced E.coli cells containing a plasmid with Pconst-luxR-terminator-Plux-rfp-terminator with different concentration of bought AHL. Upon bindung of AHL to LuxR it represses RFP prodcution. The Plux is a engineered version of the natural one, where the operon site for LuxR is placed between the -10 and -35 region of the promoter.
Copy number test
Results for BBa_K625005
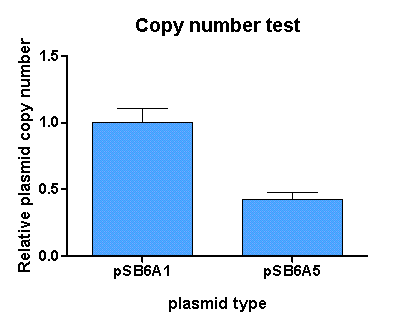
[http://partsregistry.org/Part:BBa_K625005 pSB6A5] is the version 5 BioBrick vector backbone of the medium copy (minimal pBR322 origin) pSB6A1 plasmid. This means, that the cassettes for the ORI and antibiotic resistance have been minimized (reduction of the size and transcriptional terminators flanking the prefix and the suffix have been added, to trancriptionally insulate the inserted parts from the vector backbone machinery and vice versa. This minimization yielded a reduction in size from 4022 bp to 2743 bp.
For comparison of the copy number of the plasmids, triplicates of OD normalized bacterial cultures containing the respective plasmids were miniprepped. The DNA concentration was then determined by an agarose DNA gel electrophoresis of EcoRI digests of the purified plasmids (Figure 14). This experiment showed a clear reduction in copy number of the minimized version of the original plasmid (Figure 13). Possible reasons for this could be deletion of some unknown regulatory sequences during minimization of the ORI or transcription from within the insert into the origin region in psB6A1 impacting the replication machinery.
References
[1] [http://www.pnas.org/content/97/12/6640.full.pdf+html Kirill A. Datsenko and Barry L. Wanner: One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products 2000, Proc Natl Acad Sci USA, 97: 6640-5]
[2] [http://www.ncbi.nlm.nih.gov/pubmed/3072264 Beatrice Felenbok et al. The ethanol regulon in Aspergillus nidulans: characterization and sequence of the positive regulatory gene alcR Gene 1988 ; 73(2): 385-96]
[3] [http://www.ncbi.nlm.nih.gov/pubmed/11550794 Beatrice Felenbok, Michel Flipphi and Igor Nikolaev: Ethanol Catabolism in Aspergillus nidulans: A Model System for Studying Gene Regulation, Progress in Nucleic Acid Research and Molecular Biology, 69: 149-204]
[4] [http://aem.asm.org/cgi/content/abstract/64/2/748 S. Panke, J. M. Sanchez Romero, V. de Lorenzo: Engineering of Quasi-Natural Pseudomonas putida Strains for Toluene Metabolism through an ortho-Cleavage Degradation Pathway , Applied and Environmental Microbiology, 1998, 64: 748-751]
[5] [http://www.nature.com/nature/journal/v434/n7037/full/nature03461.html Subhayu Basu, Yoram Gerchman1, Cynthia H. Collins, Frances H. Arnold & Ron Weiss: A synthetic multicellular system for programmed pattern formation, Nature 2005, 434: 1130-11342]
 "
"