Team:ETH Zurich/Biology
From 2011.igem.org
(Difference between revisions)
(→BioBricks and experimental results) |
(→Biology - Overview) |
||
| (21 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:ETH Zurich/Templates/ | + | {{:Team:ETH Zurich/Templates/HeaderNew}} |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | {| | + | {{:Team:ETH Zurich/Templates/SectionStart}} |
| - | + | = Biology - Overview = | |
| + | '''A brief overview of our SmoColi system can be found here, click on the experiments or subparts to jump to the corresponding sections.''' | ||
| + | {{:Team:ETH Zurich/Templates/SectionEnd}} | ||
| + | |||
| + | {{:Team:ETH Zurich/Templates/SlideShow|Items=<html> | ||
| + | |||
| + | <div><img src="/wiki/images/d/d9/ETH-SLIDESHOW_BIOLOGY-1.PNG"/></div> | ||
| + | |||
| + | <div><img src="/wiki/images/2/21/ETH-SLIDESHOW_BIOLOGY1.PNG" usemap="#BIOSlide1"/> | ||
| + | <map name="BIOSlide1"> | ||
| + | <area shape="rect" alt="" title="" coords="182,86,436,128" href="/Team:ETH_Zurich/Biology/Detector#Acetaldehyde_Sensor"/> | ||
| + | <area shape="rect" alt="" title="" coords="188,144,442,186" href="/Team:ETH_Zurich/Biology/Detector#Xylene_Sensor" target="" /> | ||
| + | <area shape="rect" alt="" title="" coords="188,196,442,238" href="/Team:ETH_Zurich/Biology/Detector#Arabinose_Sensor" target="" /> | ||
| + | <area shape="rect" alt="" title="" coords="72,244,638,338" href="/Team:ETH_Zurich/Biology/Detector#Bandpass" target="" /> | ||
| + | <area shape="rect" alt="" title="" coords="72,342,444,436" href="/Team:ETH_Zurich/Biology/Detector#Quorum_Sensing_of_the_Alarm_system" target="" /> | ||
| + | <area shape="rect" alt="" title="" coords="458,92,626,132" href="/Team:ETH_Zurich/Biology/Validation#AlcR-_the_acetaldehyde_sensor" target="" /> | ||
| + | <area shape="rect" alt="" title="" coords="456,148,624,188" href="/Team:ETH_Zurich/Biology/Validation#XylR_-_the_xylene_sensor" target="" /> | ||
| + | <area shape="rect" alt="" title="" coords="454,198,622,238" href="/Team:ETH_Zurich/Biology/Validation#AraC_-_the_arabinose_sensor" target="" /> | ||
| + | <area shape="rect" alt="" title="" coords="456,348,624,390" href="/Team:ETH_Zurich/Biology/Validation#Alarm" /> | ||
| + | <area shape="rect" alt="" title="" coords="638,24,748,446" href="/Team:ETH_Zurich/Biology/Validation" target="" /> | ||
| + | |||
| + | </map> | ||
| + | |||
| + | </div> | ||
| + | <div><img src="/wiki/images/8/8d/ETH-SLIDESHOW_BIOLOGY2.PNG" usemap="#BIOSlide2" /> | ||
| + | <map name="BIOSlide2"> | ||
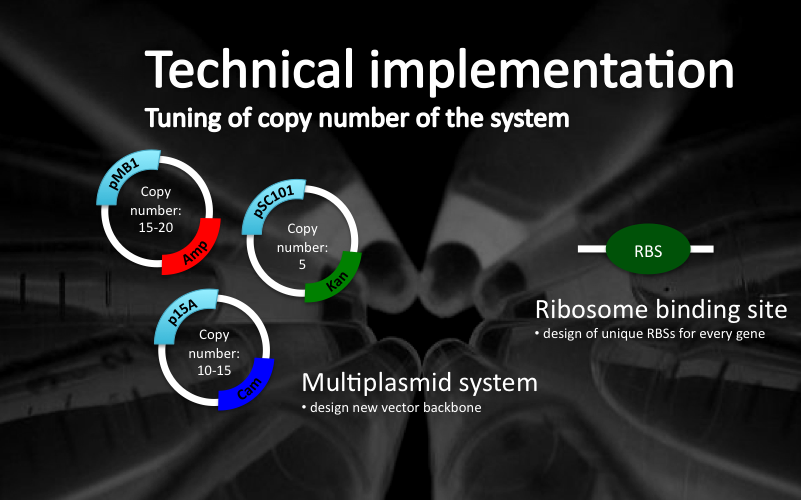
| + | <area shape="rect" alt="" title="" coords="38,124,494,412" href="/Team:ETH_Zurich/Biology/MolecularMechanism#Plasmid_design" /> | ||
| + | <area shape="rect" alt="" title="" coords="502,142,740,378" href="/Team:ETH_Zurich/Biology/MolecularMechanism#Definition_of_ribosome_binding_sites" target="" /> | ||
| + | </map> | ||
| + | </div> </html> | ||
| + | }} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--n | ||
[[File:ETH SmoColi.png|200px|left|thumb|'''Overview of the SmoColi system.''' Depending on the concentration of smoke respectively sensor molecule, a GFP band appears at a certain distance from the entry of the tube. Beyond a certain concentration cells turn red.]] | [[File:ETH SmoColi.png|200px|left|thumb|'''Overview of the SmoColi system.''' Depending on the concentration of smoke respectively sensor molecule, a GFP band appears at a certain distance from the entry of the tube. Beyond a certain concentration cells turn red.]] | ||
| Line 36: | Line 60: | ||
| | | | ||
[[File:SmoColi.JPG|350px|right|thumb|'''SmoColi pixels''' | [[File:SmoColi.JPG|350px|right|thumb|'''SmoColi pixels''' | ||
| - | :)As a side project we | + | :)As a side project we sorted and spotted ''E. coli'' expressing either RFP, GFP or β-galactosidase with a FACS machine.]] |
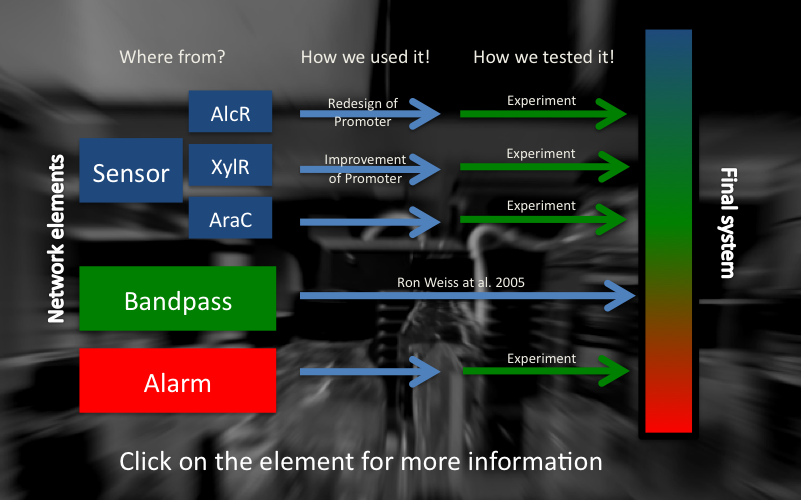
== Network elements == | == Network elements == | ||
| Line 48: | Line 72: | ||
3. alarm system | 3. alarm system | ||
| - | Both a repressor (coupled to an inverter) or an activator can be used as sensor parts of the system. In our case we used AlcR (from '' | + | Both a repressor (coupled to an inverter) or an activator can be used as sensor parts of the system. In our case we used AlcR (from ''Aspergillus nidulans''), respectively XylR (from ''Pseudomonas putida'') regulatory proteins in order to sense compounds present in smoke and thereby to induce the different parts of our network. |
Upon sensing the specific molecule the circuit gets activated and our SmoColi bacteria start notifying us about the presence of toxic substances in air. For a detailed description of the circuit design please [https://2011.igem.org/Team:ETH_Zurich/Biology/Detector click here]. | Upon sensing the specific molecule the circuit gets activated and our SmoColi bacteria start notifying us about the presence of toxic substances in air. For a detailed description of the circuit design please [https://2011.igem.org/Team:ETH_Zurich/Biology/Detector click here]. | ||
| - | + | |} --> | |
| - | + | {{:Team:ETH Zurich/Templates/HeaderNewEnd}} | |
| - | + | ||
Latest revision as of 20:50, 28 October 2011
 "
"