Team:EPF-Lausanne/Notebook/June2011
From 2011.igem.org
(→Thursday, 30 June 2011) |
|||
| Line 197: | Line 197: | ||
== Thursday, 30 June 2011 == | == Thursday, 30 June 2011 == | ||
| - | Alina and Lilia did MITOMI on TetR-GFP expressed from plasmid, it was loaded on chip in ITT expression mix (Promega TNT® SP6 High-Yield Wheat Germ Protein Expression System | + | Alina and Lilia did MITOMI on TetR-GFP expressed from plasmid, it was loaded on chip in ITT expression mix (Promega TNT® SP6 High-Yield Wheat Germ Protein Expression System). |
| - | ). DNA was spotted on June 29 in different concentrations for both: consensus (tetO1) sequence and a random sequence ((-)control). | + | DNA was spotted on June 29 in different concentrations for both: consensus (tetO1) sequence and a random sequence ((-)control). |
[[File:MITOMI_30juin_tetR_consensus-negctrl_check.png|600px|GFP-tetR and cy5 fluorescence]] | [[File:MITOMI_30juin_tetR_consensus-negctrl_check.png|600px|GFP-tetR and cy5 fluorescence]] | ||
| Line 208: | Line 208: | ||
{{:Team:EPF-Lausanne/Templates/Footer}} | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
| - | |||
== Friday, 1st of July 2011== | == Friday, 1st of July 2011== | ||
Revision as of 15:44, 1 July 2011
Notebook: June 2011
Contents |
Friday, 3 June 2011
Lysis device sequences were received from Microsynth. Only two reactions worked: those containing primers 1289F and 1333R.
Output (For the reverse primers, the reverse complement sequence is shown):
>1556291 30F
NNNNN
>1556292 440F
NNNNN
>1556293 947F
NNNNN
>1556294 1289F
NNNCTTCGGNGGGCCTTTCTGCGTTNNNATACTANTAGCGGCCGCTGCAGTCCGGCAAAAAAGGGCAAGGTGTCACCACCCTGCCCTTTTTCTTTAAAAC
CGAAAAGATTACTTCGCGTTATGCAGGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGT
AATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCG
TTTTTCCACAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTC
CCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAG
CTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGT
AACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTA
CAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAANAAGAGTTGG
TAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAANAAGGATCTCAAGAAGATCCT
TTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAANACTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTT
TAAATTAAAAATGAAGTTTTANATCAATCTAAAGTATATATGANTAAACTTGGTCTGANAGCTCGAGATTCTCATGTTTGANAGCTTATCATCGATAAGC
TTTAATGNGGTAGTTTATCNNAGTTAAATTGCTAACGCAGTCAGGNNCCGNN
>1556295 816R
NNNNN
>1556296 1333R
TNCNTTNCTAGAGGGNCAAGCAATGCTTGCCTTGAATANNAACTTTTGAATANNGATTCAGGAGGTACTAGAATGGCTTCCAAGGTGTACGACCCCGAGC
AACGCAAACGCATGATCACTGGGCCTCAGTGGTGGGCTCGCTGCAAGCAAATGAACGTGCTGGACTCCTTCATCAACTACTATGATTCCGAGAAGCACGC
CGAGAACGCCGTGATTTTTNTGCATGGTAACGCTGCCTCCAGCTACCTGTGGAGGCACGTCGTGCCTCACATCGAGCCCGTGGCTAGATGCATCATCCCT
GATCTGATCGGAATGGGTAAGTCCGGCAAGAGCGGGAATGGCTCATATCGCCTCCTGGATCACTACAAGTACCTCACCGCTTGGTTCGAGCTGCTGAACC
TTCCAAAGAAAATCATCTTTGTGGGCCACGACTGGGGGGCTTGTCTGGCCTTTCACTACTCCTACGAGCACCAAGACAAGATCAAGGCCATCGTCCATGC
TGAGAGTGTCGTGGACGTGATCGAGTCCTGGGACGAGTGGCCTGACATCGAGGAGGATATCGCCCTGATCAAGAGCGAAGAGGGCGAGAAAATGGTGCTT
GAGAATAACTTCTTCGTCGAGACCATGCTCCCAAGCAAGATCATGCGGAAACTGGAGCCTGAGGAGTTCGCTGCCTACCTGGAGCCATTCAAGGAGAAGG
GCGAGGTTAGACGGCCTACCCTCTCCTGGCCTCGCGAGATCCCTCTCGTTAAGGGAGGCAAGCCCGACGTCGTCCAGATTGTCCGCAACTACAACGCCTA
CCTTCGGGCCAGCGACGATCTGCCTAAGATGTTCATCGAGTCCGACCCTGGGTTCTTTTCCAACGCTATTGTCGAGGGAGCTAAGAAGTTCCCTAACACC
GAGTTCGTGAAGGTGAAGGGCCTCCACTTCAGCCAGGAGGACGCTCCAGATGAAATGGGTAAGTACATCAAGAGCTTCGTGGAGCGCGTGCTGAAGANNN
GAGCAGTAAACTAGAGCCAGGCATCAAATAAAACGAAAGGNTCAGTCGAAAGACTGGGCCTTTCGTTTNNNN
>1556297 1734R
NNNNN
Monday, 6 June 2011
Irina and Nadine selected the 2 plasmid backbones we will use: J23019 and J61002 (check the parts registry). They have 2 different origin of replication sites and should both have 15-20 copy numbers. Interesting also, they both contain RFP and pTet. We already have them in the freezers, now let's focus on primers design!
Wednesday, 8 June 2011
Lilia and Douglas miniprep'ed the cultures containing the lysis cassette, and sent the plasmids for sequencing.
Three colonies were miniprep'ed: two containing the T4 variant, one containing the lambda variant. This yielded solutions with plasmid concentrations of 221.9 ng/µl for T4 colony 1, 225.5 ng/µl for T4 colony 2, and 57.0 ng/µl for lambda colony. Only T4 colony 1 was sent for sequencing, using the primer solutions prepared on 31 May 2011. Results should be in on Friday.
Friday, 10 June 2011
Sequencing results from Microsynth for the lysis cassette (Biobrick K112808) have arrived. The sequence is entirely covered by the seven reactions, and all but a few bases match. As far as I can tell, this biobrick is valid! (Doug)
The results of a blast, individually querying each of the sequences against the official K112808 sequence are available here: BLAST results.
Sequencing results, for each primer (again, reverse complements shown here for the reverse primers --- those denoted by an "R"):
>1558332 30F NGNNNTCATTTGGTGTTCAGATCGCTTGTTCAAGATAACGCTACCGGGAAGGTTCTTGCTTCCCGGGTAGCTGTCGTAATTCTTTTGTTTATAATGGCGA TTGTTTGGTATAGGGGAGATAGTTTCTTTGAGTACTATAAGCAATCAAAGTATGAAACATACAGTGAAATTATTGAAAAGGAAAGAACTGCACGCTTTGA ATCTGTCGCCCTGGAACAACTCCAGATAGTTCATATATCATCTGAGGCAGACTTTAGTGCGGTGTATTCTTTCCGCCCTAAAAACTTAAACTATTTTGTT GATATTATAGCATACGAAGGAAAATTACCTTCAACAATAAGTGAAAAATCACTTGGAGGATATCCTGTTGATAAAACTATGGATGAATATACAGTTCATT TAAATGGACGTCATTATTATTCCAACTCAAAATTTGCTTTTTTACCAACTAAAAAGCCTACTCCCGAAATAAACTACATGTACAGTTGTCCATATTTTAA TTTGGATAATATCTATGCTGGAACGATAACCATGTACTGGTATAGAAATGATCATATAAGTAATGACCGCCTTGAATCAATATGTGCTCAGGCGGCCAGA ATATTAGGAAGGGCTAAATAATTAACTAGAATACTTAGGAGGTATTATGAATATATTTGAAATGTTACGTATAGATGAAGGTCTTAGACTTAAAATCTAT AAAGACACAGAAGGCTATTACACTATTGGCATCGGTCATTTGCTTACAAAAAGTCCATCACTTAATGCTGCTAAATCTGAATTAGATAAAGCTATTGGGC GTAATTGCAATGGTGTAATTACAAAAGATGAGGCTGAAAAACTCTTTAATCAGGATGTTGATGCTGCTGTTCGCGGAATCCTGAGAAATGCTAAATTAAA ACCGGTTTATGATTCTCTTGATGCGGTTCGTCGCTGTGCATTGATTAATATGGTTTTCCAAATGGGANAAACCGGTGTGGCAGGATTTACTAACTCTTTA CGTATGCTTCAACAAAAACGCTGGGATGAAGCAGCAGTTAACTTAGCTAANAGTAGATGGNNTAATCAAACNCCTAATCNNNNAAAACGAGTCATTACAA CGTTTANAACTGGNNN >1558333 440F NNNATTCCNACTCAAATTTGCTTTTTTACCAACTAAAAAGCCTACTCCCGAAATAAACTACATGTACAGTTGTCCATATTTTAATTTGGATAATATCTAT GCTGGAACGATAACCATGTACTGGTATAGAAATGATCATATAAGTAATGACCGCCTTGAATCAATATGTGCTCAGGCGGCCAGAATATTAGGAAGGGCTA AATAATTAACTAGAATACTTAGGAGGTATTATGAATATATTTGAAATGTTACGTATAGATGAAGGTCTTAGACTTAAAATCTATAAAGACACAGAAGGCT ATTACACTATTGGCATCGGTCATTTGCTTACAAAAAGTCCATCACTTAATGCTGCTAAATCTGAATTAGATAAAGCTATTGGGCGTAATTGCAATGGTGT AATTACAAAAGATGAGGCTGAAAAACTCTTTAATCAGGATGTTGATGCTGCTGTTCGCGGAATCCTGAGAAATGCTAAATTAAAACCGGTTTATGATTCT CTTGATGCGGTTCGTCGCTGTGCATTGATTAATATGGTTTTCCAAATGGGAGAAACCGGTGTGGCAGGATTTACTAACTCTTTACGTATGCTTCAACAAA AACGCTGGGATGAAGCAGCAGTTAACTTAGCTAAAAGTAGATGGTATAATCAAACACCTAATCGCGCAAAACGAGTCATTACAACGTTTAGAACTGGCAC TTGGGACGCGTATAANAATCTATAAAGCACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTG TCGGTGAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGAGTTGACAGCTAGCTCAGTCCTAGGGACTA TGCTAGCTACTAGAGATAGGAGGCCTTTATGGCCTTAAAAGCAACAGCACTTTTTGCCATGCTAGGATTGTCATTTGTTTTATCTCCATCGATTGAAGCG AATGTCNATCCTCATTTTGATAAATTTATGGAATCTGGNNTTAGGCACGTTTATNTGCTTTTTGAAAANAAAANCGTANAATCNNCTGAACAATTCTATA GTTTTANNNNAACGACCTATAAAANTGACCCNNGN >1558334 947F NNTTGNNGCGGTTCGTCGCTGTGCATTGATTAATATGGTTTTCCAAATGGGAGAAACCGGTGTGGCAGGATTTACTAACTCTTTACGTATGCTTCAACAA AAACGCTGGGATGAAGCAGCAGTTAACTTAGCTAAAAGTAGATGGTATAATCAAACACCTAATCGCGCAAAACGAGTCATTACAACGTTTAGAACTGGCA CTTGGGACGCGTATAAAAATCTATAAAGCACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTT GTCGGTGAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGAGTTGACAGCTAGCTCAGTCCTAGGGACT ATGCTAGCTACTAGAGATAGGAGGCCTTTATGGCCTTAAAAGCAACAGCACTTTTTGCCATGCTAGGATTGTCATTTGTTTTATCTCCATCGATTGAAGC GAATGTCGATCCTCATTTTGATAAATTTATGGAATCTGGTATTAGGCACGTTTATATGCTTTTTGAAAATAAAAGCGTAGAATCGTCTGAACAATTCTAT AGTTTTATGAGAACGACCTATAAAAATGACCCGTGCTCTTCTGATTTTGAATGTATAGAGCGAGGCGCGGAGATGGCACAATCATACGCTAGAATTATGA ACATTAAATTGGAGACTGAATGANATACTAGAGCCGGCTTATCGGTCAGTTTCACCTGATTTACGTAAAAACCCGCTTCGGCGGGTTTTTGCTTTTGGAG GGGCAGAAAGATGAATGACTGTCCACGACGCTATACCCAAAAGAAATACTAGTCTGCAGGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCT GCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAG GCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCNCAGGCTCCGCCCCCCTGACGANCATCNCAAAAATCGACGCTCAAGTCANANGNN >1558335 1289F NNCTTCGGNGGGCTTTCTGNGTTTATATANTAGAGTTGACAGCTAGCTCAGTCCTAGGGACTATGCTAGCTACTAGAGATAGGAGGCCTTTATGGCCTTA AAAGCAACAGCACTTTTTGCCATGCTAGGATTGTCATTTGTTTTATCTCCATCGATTGAAGCGAATGTCGATCCTCATTTTGATAAATTTATGGAATCTG GTATTAGGCACGTTTATATGCTTTTTGAAAATAAAAGCGTAGAATCGTCTGAACAATTCTATAGTTTTATGAGAACGACCTATAAAAATGACCCGTGCTC TTCTGATTTTGAATGTATAGAGCGAGGCGCGGAGATGGCACAATCATACGCTAGAATTATGAACATTAAATTGGAGACTGAATGAAATACTAGAGCCGGC TTATCGGTCAGTTTCACCTGATTTACGTAAAAACCCGCTTCGGCGGGTTTTTGCTTTTGGAGGGGCAGAAAGATGAATGACTGTCCACGACGCTATACCC AAAAGAAATACTAGTCTGCAGGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATAC GGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTT CCACAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCT GGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCAC GCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTA TCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAANAGGATTAGCANNNNNNGGTATGTAGGNNNNNCTACAGAN TTCTTGAAGTGGNGGCCTAACTACGNNTAN >1558336 816R NTTCAGNNTNTTTTACTTTCACCAGCGTTTNTGGGNGANNAAAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGNGACACGGAAATGTTGAAT ACTCATACTCTTCCTTTTTCAATATTATTGAAGCATTTNTCAGGGTTATTGTNTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATA GNGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTGACGTCTAAGAAACCATTATTATCATGACATTAACCTATAAAAATAGGCGTATCACGAGGCAGA ATNTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGACTTAAAAGGAGGGTCTATGGCAGCACCTAGAAT ATCATNTTCGCCCTCTGATATTCTATTTGGTGTTCTAGATCGCTTGTTCAAAGATAACGCTACCGGGAAGGTTCTTGCTTCCCGGGTAGCTGTCGTAATT CTTTTGTTTATAATGGCGATTGTTTGGTATAGGGGAGATAGTTTCTTTGAGTACTATAAGCAATCAAAGTATGAAACATACAGTGAAATTATTGAAAAGG AAAGAACTGCACGCTTTGAATCTGTCGCCCTGGAACAACTCCAGATAGTTCATATATCATCTGAGGCAGACTTTAGTGCGGTGTATTCTTTCCGCCCTAA AAACTTAAACTATTTTGTTGATATTATAGCATACGAAGGAAAATTACCTTCAACAATAAGTGAAAAATCACTTGGAGGATATCCTGTTGATAAAACTATG GATGAATATACAGTTCATTTAAATGGACGTCATTATTATTCCAACTCAAAATTTGCTTTTTTACCAACTAAAAAGCCTACTCCCGAAATAAACTACATGT ACAGTTGTCCATATTTTAATTTGGATAATATCTATGCTGGAACGATAACCATGTACTGGTATAGAAATGATCATATAAGTAATGACCGCCTTGAATCAAT ATGTGCTCAGGCGGCCAGAATATTAGGAAGGGCTAAATAATTAACTAGAATACTTAGGAGGTATTATGAATATATTTGAAATGTTACGTATAGATNAAGG TCTAGACTAAANNCN >1558337 1333R CCGGGAAGGTTNTTNNNTCCCGGGTAGNNNNCGTAATNNTTNNNTNNNAATGNNGATTGTTTGTNTAGGGGAGATAGTTTNTTTGAGTACNATAAGCAAT CAAAGTNTGAAACATCCAGNGAAATTATTGAAAAGGAAANAACTGCACNCTTTGAATCTNTNGCCCTGGAACAACTCCAGATAGTTCATATNTCATNTGA GGCAGACTTTAGTGCGGTGTATTCTNTCCGCCCTAAAAACTTAAACTATTTTGTTGATATTATAGCATACGAAGGAAAATTACCTTCAACAATAAGTGAA AAATCACTTGGAGGATATCCTGTTGATAAAACTATGGATGAATATACAGTTCATTTAAATGGACGTCATTATTATTCCAACTCAAAATTTGCTTTTTTAC CAACTAAAAAGCCTACTCCCGAAATAAACTACATGTACAGTTGTCCATATTTTAATTTGGATAATATCTATGCTGGAACGATAACCATGTACTGGTATAG AAATGATCATATAAGTAATGACCGCCTTGAATCAATATGTGCTCAGGCGGCCAGAATATTAGGAAGGGCTAAATAATTAACTAGAATACTTAGGAGGTAT TATGAATATATTTGAAATGTTACGTATAGATGAAGGTCTTAGACTTAAAATCTATAAAGACACAGAAGGCTATTACACTATTGGCATCGGTCATTTGCTT ACAAAAAGTCCATCACTTAATGCTGCTAAATCTGAATTAGATAAAGCTATTGGGCGTAATTGCAATGGTGTAATTACAAAAGATGAGGCTGAAAAACTCT TTAATCAGGATGTTGATGCTGCTGTTCGCGGAATCCTGAGAAATGCTAAATTAAAACCGGTTTATGATTCTCTTGATGCGGTTCGTCGCTGTGCATTGAT TAATATGGTTTTCCAAATGGGAGAAACCGGTGTGGCAGGATTTACTAACTCTTTACGTATGCTTCAACAAAAACGCTGGGATGAAGCAGCAGTTAACTTA GCTAAAAGTAGATGGTATAATCAAACACCTAATCGCGCAAAACGAGTCATTACAACGTTTAGAACTGGCACTTGGGACGCGTATAAAAATCTATAAAGCA CTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCNTTCGNTTNN >1558338 1734R TCCAACTCAAAATTTNNTTTNTTNCCAACTAAAAANNCTNNTCCCGAAATAAACTNNANGNNNAGTNNTCCANNTNTTAATTTGGATAANNNNTNNGCTG GAACGATAACCATGTACTGGTATAGAAATGATCANNNAAGTAATGACCNCCTTGAATCAATATGTGNTCAGGNGNCCAGAATATTAGGAAGGGCTAAATA ATTAACTAGAATACTTAGGAGGTATTATGAATATATTTGAAATGTTACGTATAGATGAAGGTNTTAGACTTAAAATCTATAAAGACACAGAAGGCTATTA CACTATTGGCATCGGTCATTTGCTTACAAAAAGTCCATCACTTAATGCTGCTAAATCTGAATTAGATAAAGCTATTGGGCGTAATTGCAATGGTGTAATT ACAAAAGATGAGGCTGAAAAACTCTTTAATCAGGATGTTGATGCTGCTGTTCGCGGAATCCTGAGAAATGCTAAATTAAAACCGGTTTATGATTCTCTTG ATGCGGTTCGTCGCTGTGCATTGATTAATATGGTTTTCCAAATGGGAGAAACCGGTGTGGCAGGATTTACTAACTCTTTACGTATGCTTCAACAAAAACG CTGGGATGAAGCAGCAGTTAACTTAGCTAAAAGTAGATGGTATAATCAAACACCTAATCGCGCAAAACGAGTCATTACAACGTTTAGAACTGGCACTTGG GACGCGTATAAAAATCTATAAAGCACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGG TGAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGAGTTGACAGCTAGCTCAGTCCTAGGGACTATGCT AGCTACTAGAGATAGGAGGCCTTTATGGCCTTAAAAGCAACAGCACTTTTTGCCATGCTAGGATTGTCATTTGTTTTATCTCCATCGATTGAAGCGAATG TCGATCCTCATTTTGATAAATTTATGGAATCTGGTATTAGGCACGTTTATATGCTTTTTGAAAATAAAAGCGTAGAATCGTCTGAACAATTCTATAGTTT TATGAGAACGACCTATAAAAATGACCCGTGCTCTTCTGATTTTGAATGTATAGAGCGAGGCGCGGAGANNNCACAN
Tuesday, 21 June 2011
With help from Henrike, Douglas checked the mistakes in the K112808 (lysis cassette) sequence. On the entire sequence, only 7 errors occurred in several reactions, and are therefore likely to be actual errors in the DNA. They occur in positions (with regard to the original sequence in the registry):
- 70: CTC codon substituded for CTA. Both code for leu, so that position is still valid
The following are missing bases:
- 677 and 684: lie in the junction between sub-bricks K112805 and K112806. Still valid.
- 1199: lies in the junction between K112806 and B0010. Still valid.
- 1387: second base of K112807, in the non-coding region. Still valid.
- 1698: lies in the junction between K112807 and B0010. Still valid.
- 1701: beginning of B0010, in the non-coding region. Still valid.
In conclusion, there is one codon substitution, and six missing bases that all lie within the junction between two bricks or the non-coding region of a brick. Experimentally, the brick does lyse cells in culture. Therefore, the sequence appears valid.
Monday, 27 June 2011
We practised MITOMI with TetR linear template and random DNA sequence + target DNA sequence. We performed ITT (in vitro transcription/translation) on TetR linear template before adding it to the chip. However, the results are disappointing: we have more DNA binding for the random sequence (1000 RFU change) than for the target sequence (500 RFU)...
Images: Green fluorescence comes from TetR (labeled Lysines) and red fluorescence comes from DNA (cy-5 labeled). We can clearly see that the protein amount seems to be the same, whereas the DNA fluorescence is decreased.
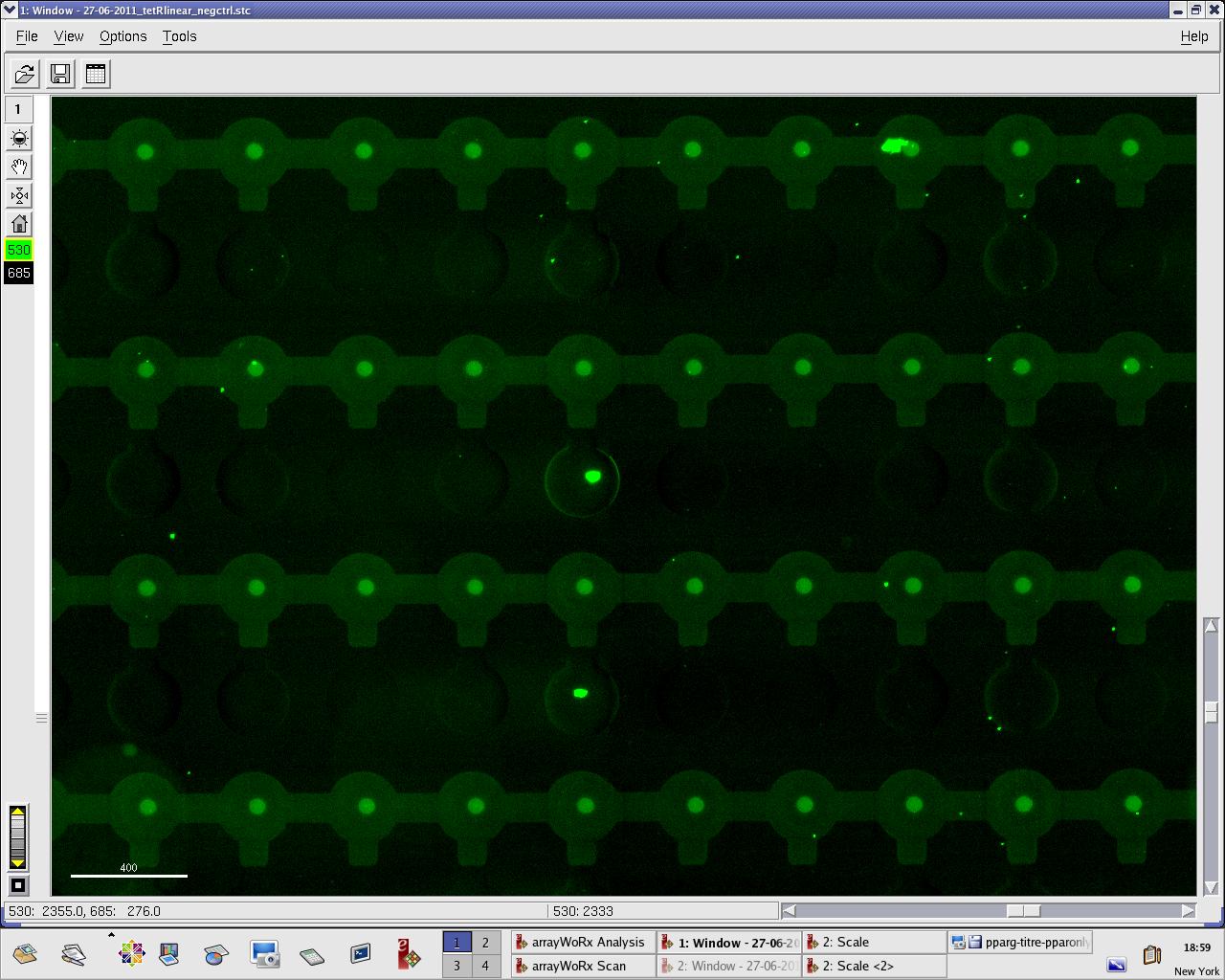
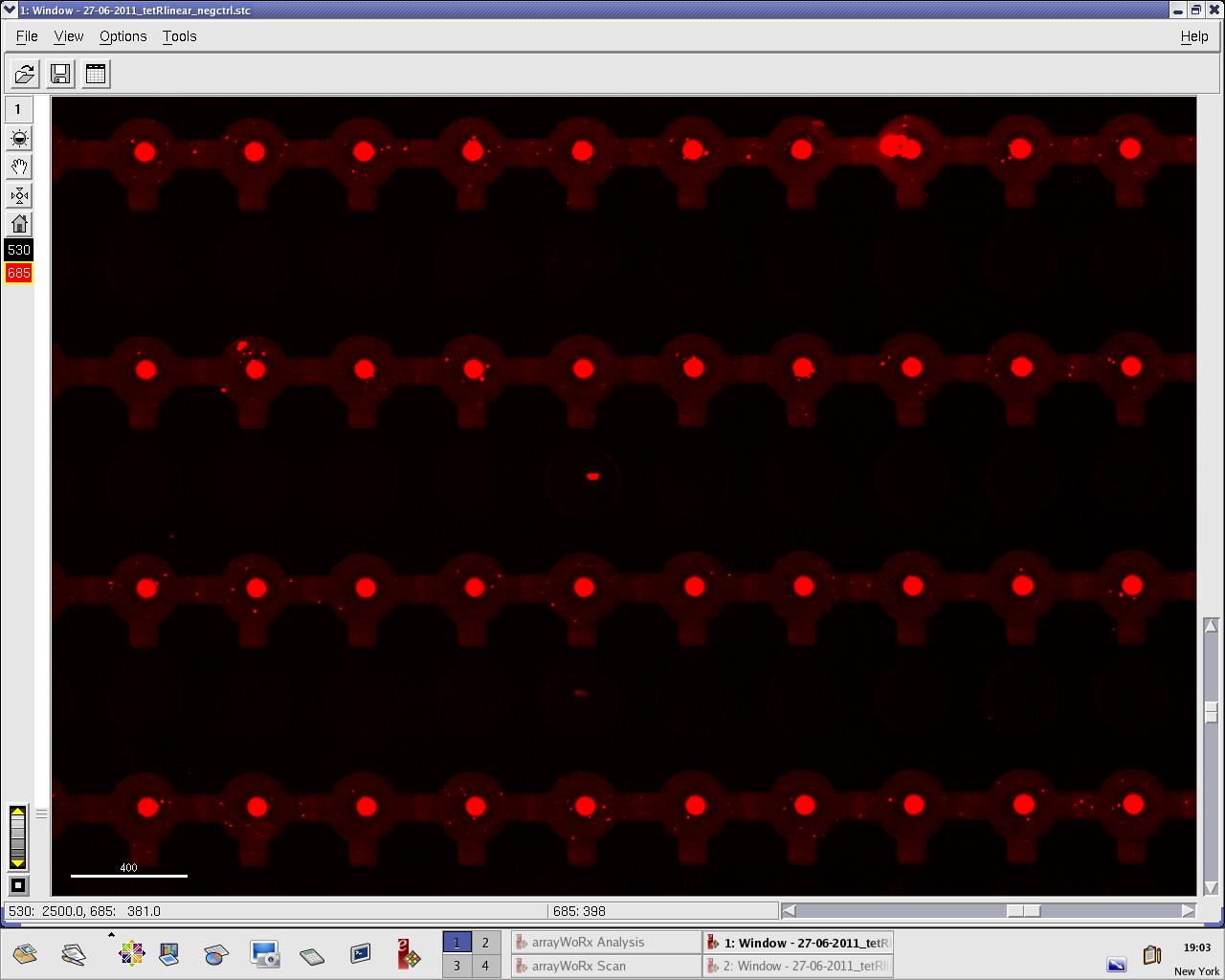
Perhaps the ITT doesn't yield a functional TetR, or we didn't have the optimal concentration of binding DNA (too much nonspecific interactions?). We'll definitely continue the MITOMI practise to find out.
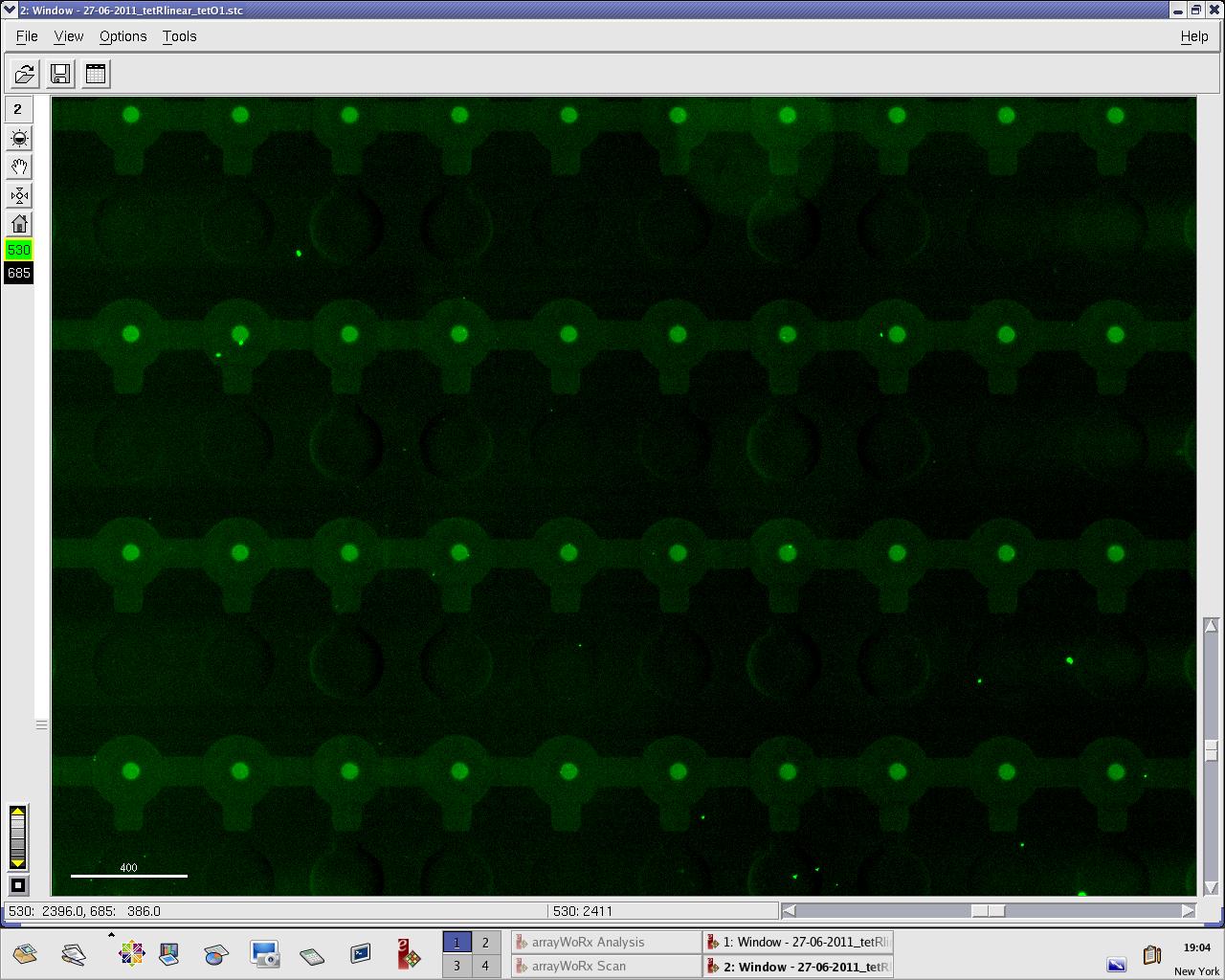
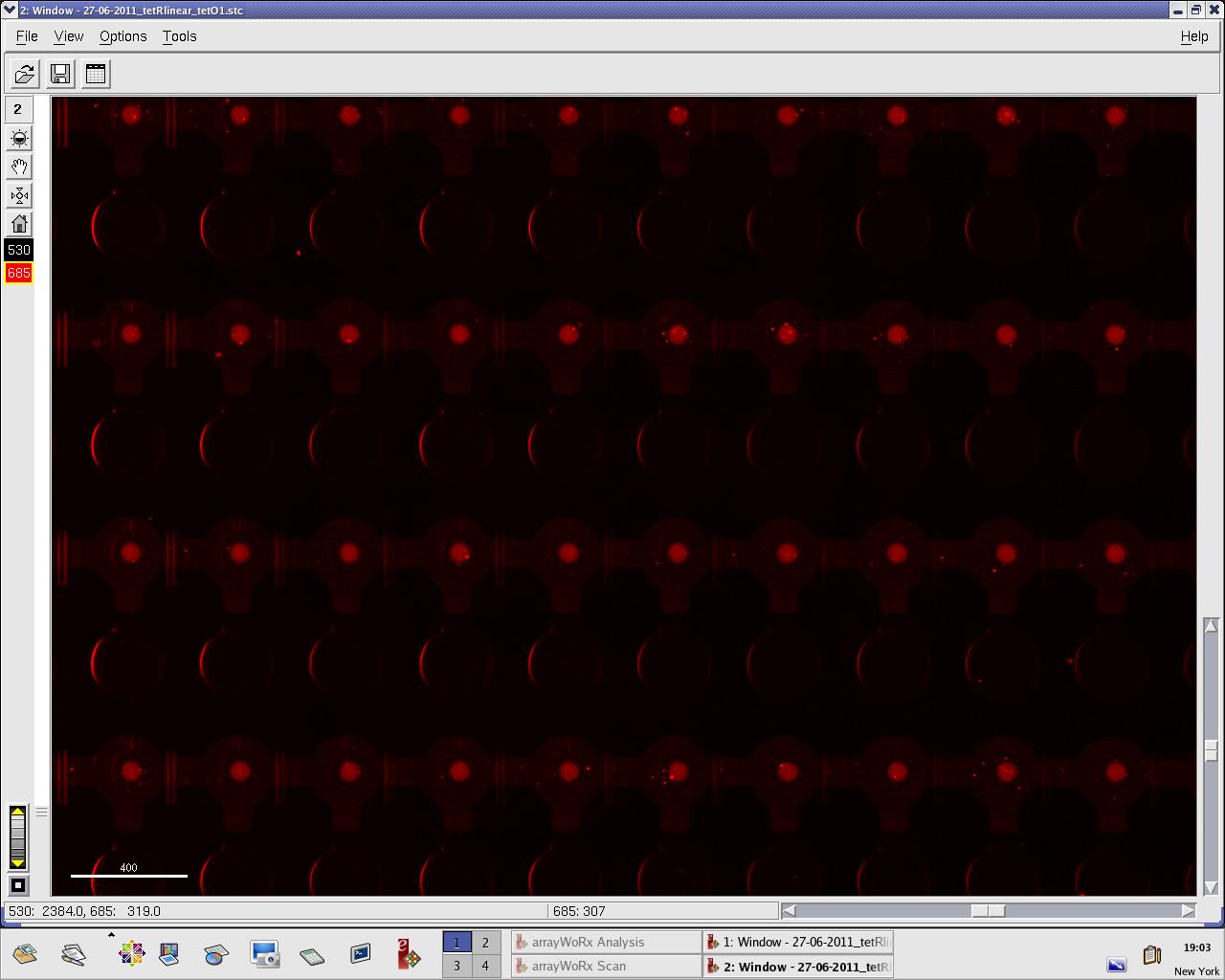
Thursday, 30 June 2011
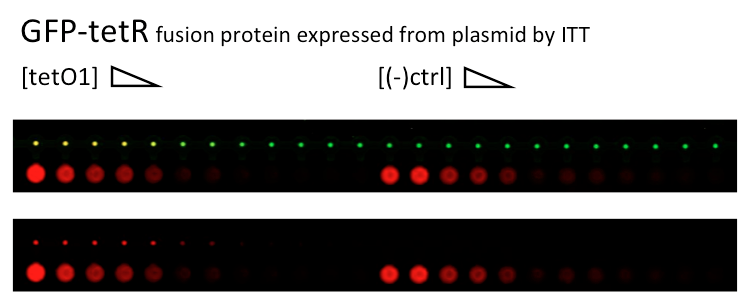
Alina and Lilia did MITOMI on TetR-GFP expressed from plasmid, it was loaded on chip in ITT expression mix (Promega TNT® SP6 High-Yield Wheat Germ Protein Expression System). DNA was spotted on June 29 in different concentrations for both: consensus (tetO1) sequence and a random sequence ((-)control).
Images: Green fluorescence comes from GFP-TetR and red fluorescence comes from DNA (cy-5 labeled).
This experiment confirms that wtTetR is expressed and correctly folded by SP6 expression system. The TF recognizes tetO1 consensus sequence while the tested random sequence (-)control has a significantly lower binding affinity to TetR.
Friday, 1st of July 2011
Alina and Lilia did MITOMI on His-TetR expressed from linear template, it was loaded on chip in ITT expression mix. DNA was spotted on June 29 in different concentrations for both: consensus (tetO1) sequence and a random sequence ((-)control).
Images: Green fluorescence comes from green-lysine and red fluorescence comes from DNA (cy-5 labeled).
Although we don't observe much protein bound to the anti His-tag antibody, we can still see that it does bound TetO1 sequence and did not bind any (-)control sequence. Some possible reasons for low protein fluorescence under the buttons: the expression yield from the linear template with T7 promoter might be too low, the His-tag to antibody binding is not strong enough or the concentration of antibody is too low.
 "
"