Team:EPF-Lausanne/Notebook/August2011
From 2011.igem.org
(→Friday, 26 August 2011) |
|||
| Line 400: | Line 400: | ||
| + | [[File:EPFL_Igem_2608_j6RFPcol24.jpg|thumb|300px]] | ||
| + | [[File:EPFL_Igem_2608_pcr13.jpg|thumb|300px]] | ||
Nadine made colony PCRs for yesterday's Gibson assemblies: J61002 Plac-RFP and pSB3K1 TetR-LacI; she also made PCRs for pSB3K1 TetR-LacI that had been sent for sequencing on wednesday. Unfortunately, the plate with J61002 Plac-lysis yielded no colonies... | Nadine made colony PCRs for yesterday's Gibson assemblies: J61002 Plac-RFP and pSB3K1 TetR-LacI; she also made PCRs for pSB3K1 TetR-LacI that had been sent for sequencing on wednesday. Unfortunately, the plate with J61002 Plac-lysis yielded no colonies... | ||
Details of the PCRs: | Details of the PCRs: | ||
| Line 408: | Line 410: | ||
** amplify RFP with colony PCR primers | ** amplify RFP with colony PCR primers | ||
** amplify RFP with Gibson primers | ** amplify RFP with Gibson primers | ||
| + | |||
With the sequencing primers, one of them binding in the middle of RFP, all 6 colony tested gave positive results. Strangely, there seems to be 2 bands with a slight difference in size. With the Gibson primers that contain Plac, colonies 1 and 3 have a brighter band. Either they are the only positive ones, or they just have a bigger amount of DNA (as also seen on the other gel). | With the sequencing primers, one of them binding in the middle of RFP, all 6 colony tested gave positive results. Strangely, there seems to be 2 bands with a slight difference in size. With the Gibson primers that contain Plac, colonies 1 and 3 have a brighter band. Either they are the only positive ones, or they just have a bigger amount of DNA (as also seen on the other gel). | ||
| Line 413: | Line 416: | ||
| + | [[File:EPFL_Igem_2608_J6RFP_dig.jpg|thumb|300px]] | ||
Nadine also digested the (supposed) J61002 Plac-RFP plasmids from Tuesday's Gibson assembly: colony 6 from Gibson plate that was red, colony 9 from Gibson plate that was white and colony 12 from negative control plate that was also red. Pst1 has a restriction site in the original J61002 backbone, therefore it should cut independently of RFP insertion. The results indicate that only colony 9 was digested by the enzyme, therefore it's likely that the red colonies contain an unknown plasmid that has RFP... Since colony 9 was positive for RFP colony PCR, it can be our expected plasimid that has little transcription of RFP. | Nadine also digested the (supposed) J61002 Plac-RFP plasmids from Tuesday's Gibson assembly: colony 6 from Gibson plate that was red, colony 9 from Gibson plate that was white and colony 12 from negative control plate that was also red. Pst1 has a restriction site in the original J61002 backbone, therefore it should cut independently of RFP insertion. The results indicate that only colony 9 was digested by the enzyme, therefore it's likely that the red colonies contain an unknown plasmid that has RFP... Since colony 9 was positive for RFP colony PCR, it can be our expected plasimid that has little transcription of RFP. | ||
{{:Team:EPF-Lausanne/Templates/Footer}} | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
Revision as of 09:29, 27 August 2011
Notebook: August 2011
Tuesday, 02 August 2011
Lilia came on Saturday to take the plates and here are the results:
The old competents cells seem not to work very well and the Gibson assembly failed since in the plate with only backbone we have a lot of cells therefore the backbone can closes on itself.
Alessandro make the PCR (gradient PCR in which the annealing temperature increases for each sample) to amplify TetR (putting Pconst in front of it) from Repressilator plasmid to make the Gibson for the Pconst-TetR plasmid. The result is illustrated on the right.
Since we have already amplified the backbone (pSB3K1) we can make the Gibson assembly to make the Pconst-TetR plasmid.
The latest mutation PCR yielded large amounts of product. The two bands might be a little worrying, but overall this is a lot better than any of the previous trials, with the old polymerase. The stitch PCR will be ran tomorrow.
On the side, more reporter and lysis plasmids were assembled (together with a control, containing only the backbone), from the previously made gibson fragments (PCR purified on 2nd August 2011).
Wednesday, 03 August 2011
Alessandro made glicerol stocks of Vincent's plasmid (J61002-Ptet-RFP) and purified the PCR product of the day before (Pconst-TetR) to make the Gibson assembly (using the already amplified backbone of pSB3K1). The Gibson assembly for the pSB3K1-Pconst-TetR (using either equimolar ratio of backbone-insert either 2 times insert) and for Reporter-RFP/Lysis has been plated.
Douglas ran the stitch PCR on the latest mutants, unfortunately yielding virtually no product. Only the PQ39LV31YM42 mutant has detectable amounts of product in the correct size, but then it also had a much larger concentration of template (or so it seems by eyeballing the mutation PCR's gel).
Likely causes:
- The common sequence DNA was lost during PCR purification (Doug accidentally used the high cut-off buffer), in which case the mutation PCR should be repeated from scratch...at least for the common sequence.
- The 3' final and 5' final primers are not suitable for the stitch step. Concentrations and heat cycles should be accurate, because they worked for the mutation PCR.
Nadine transformed and plated Doug's mutant TetR plasmids from 26.07. She prepared the X-gal and IPTG solutions needed for the control, but they still need to be filtered.
We tried a first colony PCR on J61002 Ptet-RFP cells, but nothing got amplified... Tomorrow we'll try the primers on purified plasmids to see it the problem comes from here.
Thursday, 04 August 2011
The mutant TetR plasmids plated yesterday yielded only one colony, even the mutation control didn't work. Nadine plated the rest of transformed cells, but probably the cells in the mutagenesis kit are too old to be competent.
Alessandro took plates from the day before and about 20 colonies grew for Reporter-Lysis with BL-21 and less using DH5alfa cells; for Reporter-RFP just 1 colony grew; about 10 colonies for Pconst-TetR. Alessandro screened some of them with colony PCR. The results are available only for Vincent's plasmid (our positive control) and for the pSB3K1-Pconst-TetR and they are positive (but we cannot estimate the size since the ladder seems to be very degraded). For Vincent's plasmid, together with cells Alessandro also made the PCR with the purified plasmid (to check whether the primers are good).
The following day the results for the other plates will be available and Alessandro will make co-transformation with Vincent's plasmid and with psB3K1-Pconst-TetR in order to assess the expression of RFP, controlled by TetR.
Friday, 05 August 2011
Nadine checked the 2nd plating of the mutagenesis experiment, and again we had cells only for the 4th mutagenesis(about 10 colonies). She made liquid cultures from 5 of them, the miniprep will be done tomorrow.
Colony PCRs for the Reporter-lysis plasmid have been made but they failed... so we have to start over again from Gibson assembly. Only one sample for Reporter-RFP has been tested, and the colony PCR also failed.
Alessandro and Alina made Kanamicine plates and Kanamicine+Ampicillin plates. Alessandro transformed:
- pSB3K1
- Vincent's plasmid (J61002-Ptet-RFP)
- pSB3K1 Pconst-TetR
- the old Gibson assembly for Reporter-RFP
- co-transformation with pSB3K1-Pconst-TetR and Vincent's plasmid
The transformation were performed for pSB3K1 because unexpectdely the liquid cultures for pSB3K1-Pconst-TetR (positive with colony-PCR) were red. On the registry the pSB3K1 backbone hasn't been documented to have RFP so the question why this cells were red is open. This new assembly has to be sequenced.
Monday, 08 August 2011
Lilia went on Saturday to miniprep the TetR mutants, and the concentrations are really high: from 150 ng/ul to 238 ng/ul.
Nadine made new Gibson reactions for the Reporter and Lysis plasmids: once with equimolar ratios and once with about 2x more parts than the backbone, plus a negative control containing only the backbone.
Alessandro prepared SOC medium and started liquid culture of pSB3K1 and pSB3K1-Pconst-TetR to make glycerol stocks. The cells transformed with pSB3K1 are red therefore that plasmid has RFP though not documented on the registry. A lot of colonies grew on the co-transformation plate; Alessandro will start a liquid culture the following day to test them in the plate-reader.
Vincent used the QuikChange Site-Directed Mutagenesis (SDM) Kit to try to produce TetR mutants. The pWhitescript 4.5-kb control plasmid provided by the Kit was used. The plasmid contains a stop codon (TAA) rather than a glutamine codon (CAA) in the beta-galactosidase gene of the pBluescript II SK(-) phagemid. When we transform the plasmid into XL10-gold ultracompetent cells, the resulting cells should look white on LB-ampicillin plates that have been treated with IPTG and X-gal, since the beta-galactosidase is no longer functional. However, if the mutagenesis works, there should be a point mutation that reverts the T into a C (TAA into CAA), meaning that the cells should appear blue on plates that contain IPTG and X-gal.
The protocol for Mutant Strand Synthesis (provided with the Kit) was followed meticulously. Vincent made a master mix for 5 mutants and made a separate control mixture. The DNA template concentration was 130 ng/uL (after being diluted from 160 ng/uL as used by Douglas) so we used .5 uL to get around 65 ng per mutant sample. The protocol asks for 125 ng of each primer. Since the primers all had the same 1 ug/uL concentration, we used 8 uL of each primer. The mutants were the following:
- Y42F
- V36F
- P29Q-Y42M
- P39Q-LV41
- P39K
Since the total strand is about 5 kb, we used 150 seconds at 68 C for the second segment of thermal cycling. The resulting cycling products were put in the fridge over night, since it was too late to start the digestion.
Tuesday, 09 August 2011
Plates with yesterday's Gibson assemblies were not concluent, as there were few colonies without RFP and not more than in the negative control. Alessandro and Nadine made a gradient Gibson assembly for reporter plasmid and a negative control (=only backbone), we will transform them as soon as the competent cells are ready.
The mutagenesis cycling products were digested using 2 uL of Dpn I restriction enzyme. The transformation of XL10-Gold cells was done normally. The ampicillin LB plates were melting, making beading a problem. The plates were put overnight.
Lilia run MITOMI on spotted de Brujin library with GFP-wtTetR expressed off-chip with SP6 ITT. Before the last scan, the chip was washed by mistake with opened buttons, so the interactions unfortunately might have been washed on the next images.
Wednesday, 10 August 2011
Henrike and Alessandro designed primers to put the T4 lysis cassette (BBa_K112808) under the control of the T7 promoter using as backbone: pSB3C5, pSB3K1, pSB3K5. This device will be used to test plasmid DNA recovery after lysis.
Nadine designed primers to add Ptet-LacI in the pSB3K1-Pconst-TetR palsmid, also adding a terminator before LacI. We decided not to inculde the ssrA degardation tag in LacI, to be sure that the cells won't lyse.
Since the pSB3K1 plasmid contains RFP, Nadine tried to cut it out by digesting it with XbaI and SpeI. Tomorrow we'll make a ligation, and as soon as the primers arrive we'll be able to add LacI with Gibson assembly. Ligation results show 2 bands, as expected, however they are too big...
Vincent got the plates containing the tetR mutants: only the control worked, and not spectacularly well at that. The LB plates were not very good, so a new batch of plates was made with what was left of the previous day's transformation (kept overnight in the 4 C fridge). Unlike the previous plates, the sample mutagenesis plates were not covered with IPTG and Xgal, since these mutageneses do not benefit from such treatment.
Thursday, 11 August 2011
Lilia measured the competence of DH5alpha cells prepared by Clara, Lilia and Alina according to the second (Inoue) protocol for chemically competent cells, they were pipetted on Tuesday by Clara and Lilia. On Wednesday 20µl of cells were transfected with 100, 50 and 10pg of the plasmid containing GFP-TetR. The 10pg plate is shown on the picture. Competence is 10^8 CFU/µg.
Nadine ligated the pSB3K1 Pconst-tetR that should not contain RFP anymore. She also transformed it into Henrike's and our new competent cells.
Vincent got the fresh plates out of the incubator, only to find that the P39Q-Y42M was the only mutation that had grown colonies (only 2), which we consider to be an accident given that there ought to be hundreds of colonies. After poring through the protocol yet again, we spotted a mistake in the original dilution approach. Vincent went ahead and re-ran another PCR, digestion, transformation sequence, and we will see how the plates look tomorrow. In the meantime, a few emails have been sent back and forth with Cambridge's Bill Collins over the possibility of adding modules to the current Gibthon program. Discussions are ongoing.
Alessandro searched literature for T7 variant with different strength to use them in the T7-Lysis plasmid.
Lilia run MITOMI on spotted de Brujin library with GFP-wtTetR expressed off-chip with SP6 ITT. Everything went well, but no strong interactions were seen.
Chip fabrication, spotting, alignement and the whole experiment was done by Lilia. Clara's wafers were used.
Kanamycin aliquotes of 50mg/ml in water and Ampicilin aliquotes 100mg/ml in 50% ddH2O and 50% ethanol were prepare by Lilia and put in the new box labelled "iGEM2011 Antibiotics" at -20°C in BM building.
Friday, 12 August 2011
The transformed cells with digested pSB3K1 Pconst-tetR yielded only white colonies, which indicate that RFP actually got cut out. Nadine also made a colony PCR on 10 of the colonies, because the primers include the multiple cloning site. As a control, the same PCR was made on undigested pSB3K1 Pconst-tetR plasmid. The expected size for the digested would be 1200 kb, whereas with RFP it should be around 2000 bp. But the gel gives disappointing results, with every colony and the control showing the same ~800 bp band... Liquid cultures have been made, and if they don't turn red we'll miniprep them and send to sequencing. !!Uppdate: I used the tetR plasmids for Gibson assembly, so the results are fine and show that TetR is still present in the digested plasmids.
Alessandro made the extension PCR on RPF to put it under the control of 4 different T7 promoter: T7const (almost 100%), T7lac (T7 promoter with different versions of a lac operator downstream). The gel will be run on Monday as well as the gene specific PCR for T4 lysis cassette.
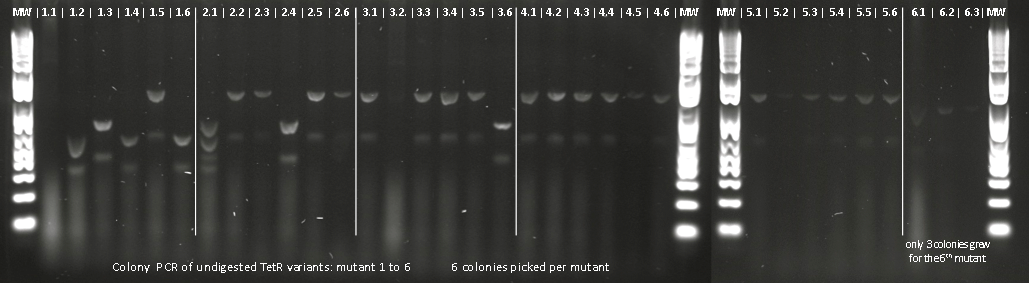
Lilia made colony PCR on TetR variants prepared by Alina and transformed on Wednesday.
Vincent took his TetR variants out of the incubator to find that 2 of the 4 had been successfully transformed and had given around 100 colonies each (the successful ones were Y42F and V36F, both single mutations). He made liquid cultures to be used for glycerol stocks and minipreps the next day. The goal is to have the variants ready for sequencing by Monday. As for the transformation efficiency, each sample reaction got 65 ng of DNA in .5 uL (eventually 50, 100, and 200 mL were plated)
Saturday, 13 August 2011
Vincent made glycerol stocks for the two mutants (Y42F, V36F -- 4 colonies each). He also miniprepped these populations with resulting concentrations being:
- Y42F 47.2 ng/uL
- V36F 31.4 ng/uL
These concentrations are low because a small amount ( 1 mL) of the liquid culture was used. If need be, the remaining 3 mL of the liquid culture can be taken out of the -4 degree fridge and used for re-doing a larger miniprep.
On the Gibthon front, Bill of Cambridge has released the most recent version of Gibthon for all to see. It has many more features than any of the previous releases, including the current version at gibthon.org (Gibthon Construct Designer). Some of these features are either features we thought we could add or are additional features that work well and enhance the program. Vincent will brainstorm this weekend about ways to further enhance the code and will present these ideas on Tuesday.
Lilia minpreped muTetR variants selected according to the colony PCR results and made glycerol stocks.
Miniprep results:
1.5 237ng/µl | 2.2 142ng/µl | 2.5 150ng/µl | 3.1 142ng/µl | 3.3 152ng/µl 4.2 64ng/µl | 4.3 140ng/µl | 5.1 69ng/µl | 5.3 65ng/µl | 6.2 44ng/µl
Nadine miniprepped the pSB3K1-pConst-TetR ligated plasmids since all the colonies were still white. She also miniprepped the J13521 (ptet-RFP) and J13522 (ptet-GFP) plasmids from registry, and made glycerol stocks for all of them.
Monday, 15 August 2011
All the LB bottles were contaminated therefore Alessandro made new ones. By now just one bottle at a time should be used by iGEMers in order to avoid multiple contamination. (take the one labeled "use me")
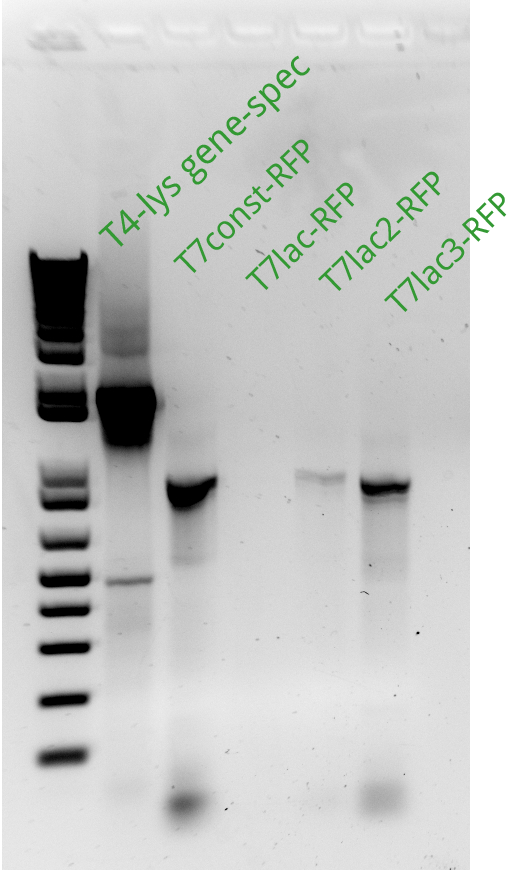
Alessandro ran the gel for the extension PCR on RFP and for gene-specific PCR on T4. In the picture there's the result: we miss a signal from T7lac-RFP and we have a weak signal from T7lac2-RFP therefore these two reactions have been repeated succesfully (see picture on the left).
Alessandro made PCRs to amplify pSB3K1 and pSB3C5 plasmids to use them as backbone for our device (T7-lysis/RFP) and they'll be ran on gel the day next. The extension PCR for T7-lysis failed because extension time was erroneously set as 1 minute instead of 2 minutes and therefore they have been repeated and will be ran on gel the following day.
Lilia run MITOMI on spotted 1-off library with GFP-wtTetR expressed off-chip with SP6 ITT. The results are being analysed.
Tuesday, 16 August 2011
Nadine got the sequencing results for digested-ligated pSB3K1 Pconst-TetR and they look fine! There is no trace of RFP, and Pconst-TetR are perfect. Seems that we have our 1st beer award. She co-transformed this plasmid with J61002 pTet-RFP and booked the plate reader, and finished ordering all the needed primers, including colony PCR primers.
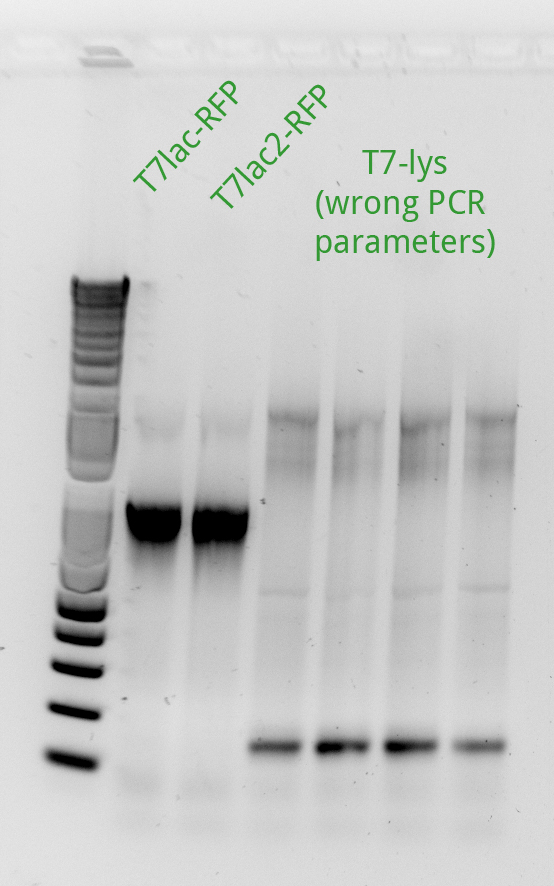
Alessandro ran the gel for the PCR products of the previous day: we miss pSB3K1 therefore the reaction has been repeated with different templates (even with the plasmid from the registry) and it worked. Another PCR has been run to amplify the backbone from pSB3K5 but it didn't work (maybe because the plasmid taken from the registry's plate has a very low concentration); the reaction will be repeated only after the miniprep for this sample will be done. Alessandro proceeded with the protocol started the day before from Henrike to make competent cells: the BL21 (DE3) which will be used for the T7-lys plasmid since these bacteria have the T7 polymerase gene under the control of Plac (they overexpress LacI). Unfortunately the PCR purification gave us bad results for the T7-lysis which is at very low concentration. pSB3C5 instead has an high concentration. Henrike suggested, to have an higher yield, to dilute your PCR products 4-fold before starting the PCR purification protocol. Alessandro PCR purified the previous succesfully amplified T7-RFP products but for T7-lac-RFP he had a very low yeld so the next day the PCR and purification has to be repeated.
Vincent got the sequences for the Y42F, V36F mutants back. With the help of the BLAST alignment tool, it became clear that the V36F mutant was as expected, while the Y42F was in fact the P39Q-Y42M mutant. It may be that the primers got mixed up or that the labelling of plates was incorrect. Either way, we have two mutants.
Wednesday, 17 August 2011
Alessandro proceeded with the day 3 of competent cells making protocol. Alessandro ran the PCR to amplify T7lac2-RFP again and it' ok (the following day he will do the PCR purification) and PCR purified the pSB3K1 backbone (succesfully amplified the day before) as well as the T7-lysis again but this time the yeld was higher. Alessandro has made the Gibson assembly for pSB3C5 and pSB3K1 with T7-RFP and with T7-Lysis (the Gibosn for the T7lac2 hasn't been done because we ran out of Gibson master mix). Alessandro miniprepped pSB3K1.
Vincent double-checked his mutant results using CLUSTALW and the results are available for File:P39q y42m official alignment clustalw.txt
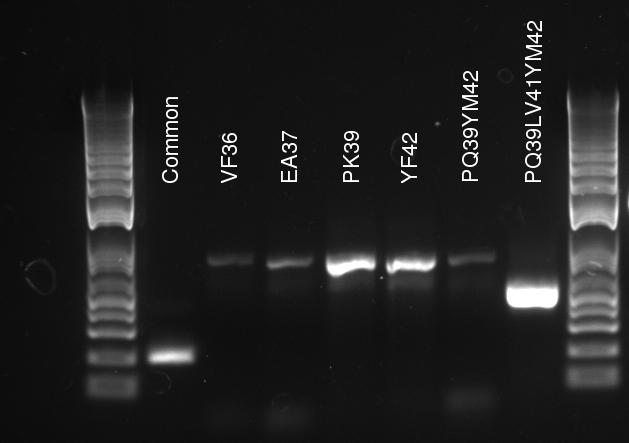
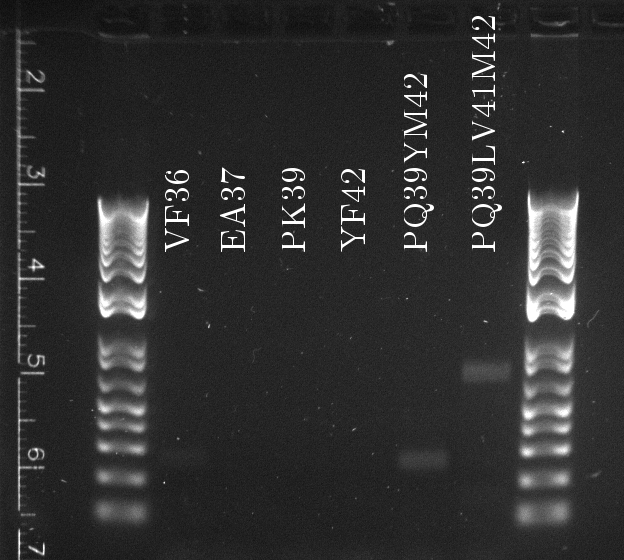
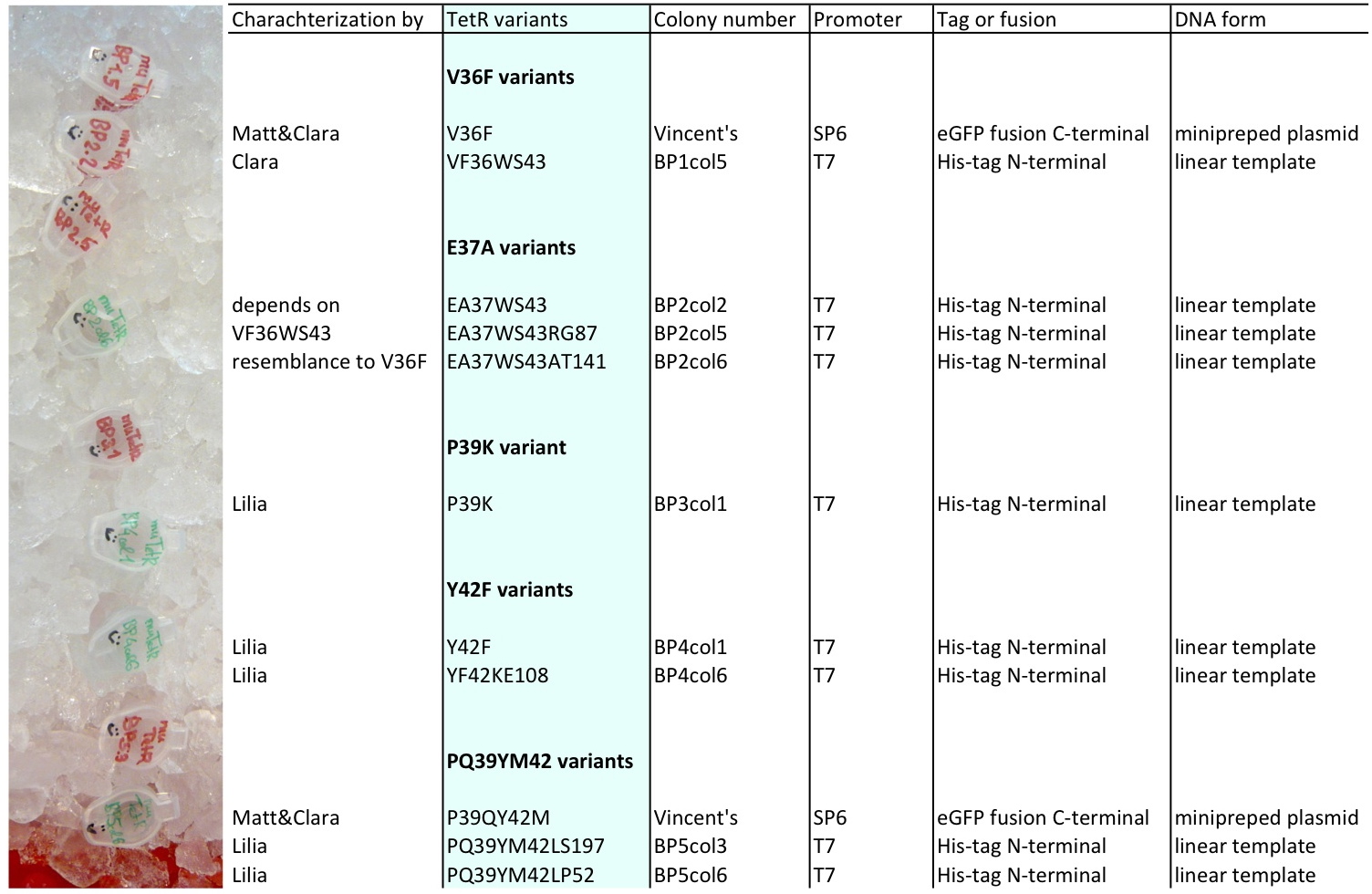
Lilia compared the received sequences of the TetR variants to the expected.
- VF36 variant: "VF36WS43"
- EA37 variants: "EA37WS43" "EA37WS43RG87"
- PK39 variant: "PK39"
- YF42 variants: "YF42SN7del" "YF42delTA112FS119"
- PQ39YM42 variants: "PQ39YM42LS197" "PQ39YM42deldel"
- PQ39LV41YM42: did not work
Nadine and Vincent made the PCR for the next Gibson assemblies:
- Amplify LacI with Ptet to put it into pSB3K1 Pconst-TetR
- Amplify pSB3K1 Pconst-TetR
- Amplify RFP with Plac to put it into J61002 plasmid
- Amplify J61002 backbone
- Amplify lsyis cassette with Plac to put it into J61002 plasmid
The parts 1 and 2 will be used to make pSB3K1 Pconst-TetR Ptet-LacI; parts 3 and 4 will yield J61002 Plac-RFP and parts 4 and 5 will yield J61002 Plac-lysis.
The PCR had faint bands, but this might come from tha fact that the gel was used for the 3rd time. Lines 1 to 4 have the expected size, but for line 5 the product is too small (it should be 1880 bp).
Thursday, 18 August 2011
Alessandro transformed the Gibson assembly of the previous day and completed the protocol for competent cells.
Nadine set the platereader experiment with the co-transformed cells containing J61002 Ptet-RFP and pSB3K1 Pconst-TetR. She ran a gradient PCR on the lysis cassette (that didn't work yesterday),but again we have a too short product... This assembly needs to be check tomorrow.
Friday, 19 August 2011
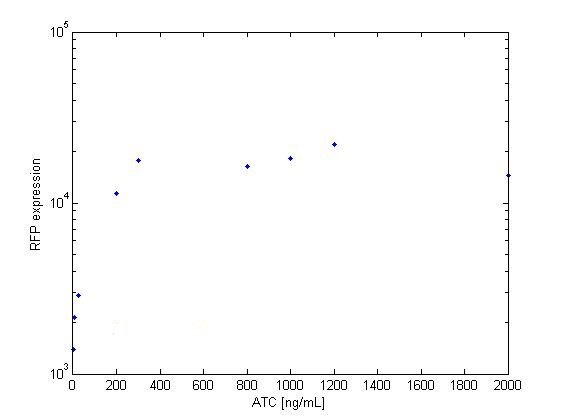
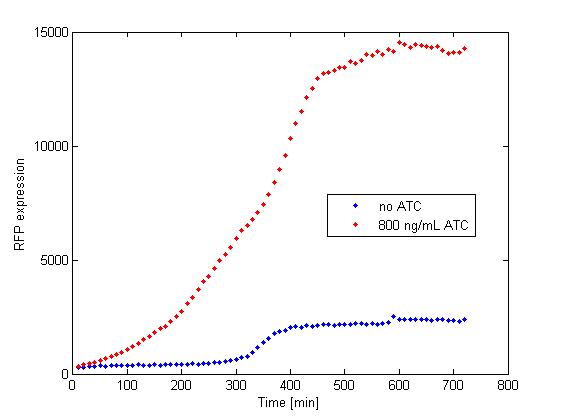
The platereader experiment worked well, showing that with enough ATC concentration RFP can be induced even with constitutive expression of TetR. The first graph shows the maximal RFP signal corresponding to each ATC dilution, averaged over the 4 different cell colonies. The second graph shows RFP expression over time for 2 individual wells, either without ATC or with 800 ng/mL. The results confirm that both pSB3K1 Pconst-TetR and J61002 Ptet-RFP plasmids work as expected.
Very high concentrations were tested, because at first we had used false dilutions that had only low impact on RFP induction. There seems to be a saturation above 300 ng/mL, so in the future we don't need to use as much high concentrations of ATC.
About the PCR of the lysis cassette for the new Gibson assembly, Nadine found out that she designed the reverse primer on a terminator (BBa_B0010) that was also present 400bp before, allowing the primer to anneal perfectly after 1400bp instead of 1800bp. This mistake was also present in the primer designed for the 3-part Gibson assembly, but we didn't notice at this time (see 11.07, the lysis plasmid has about the same size as the 1300 bp long lacI fragment). She designed a new primer, skipping the final terminator because there will be another terminator in the Gibson-assembled plasmid.
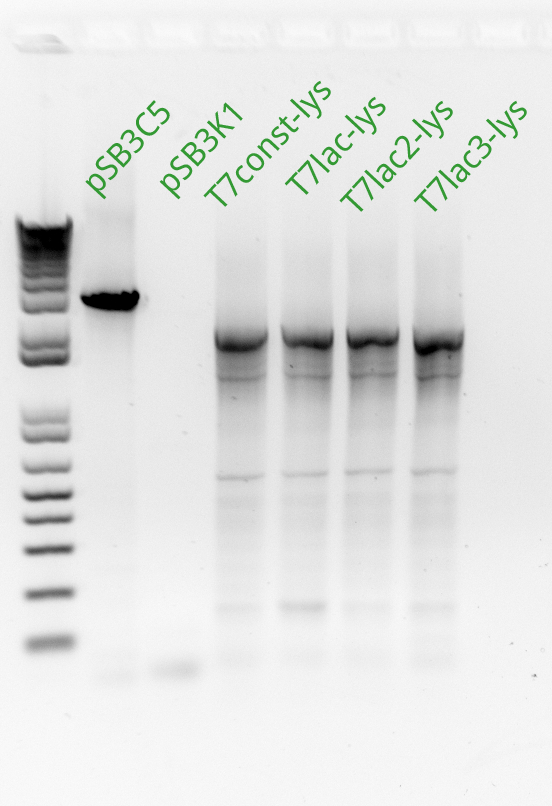
Vincent and Alessandro did a colony PCR with Taq polymerase using the K1 transformations done on Thursday. The C5 transformations yielded a small number of colonies. The gel result is below. The bright bands correspond to K1-T7-const-RFP, K1-T7-lac-FRP, K1-T7-lac2-RFP.
After noticing the poor results for the lysis, Alessandro and Vincent realized that they had set the extension PCR extension time to 1 minute rather than the 2 that are needed for the lysis cassette (which has a length of 2 kb).
Lilia compared the received sequences of the TetR variants to the expected.
- VF36 no other correct colony to test
- EA37 variant: "EA37WS43AT141"
- PK39 variant was already made: see PK39
- YF42 variants: "YF42" "YF42KE108
- PQ39YM42 variants: "PQ39YM42LP52" "PQ39YM42_ins_del"
Saturday, 20 August
As part of Friday's Colony PCR, Alessandro and Vincent had made liquid cultures and made separate, dedicated plates for each colony that was used. Since the Colony PCR had successfully amplified the K1-T7-RFP plasmids, Vincent brought out those liquid cultures, made glycerol stocks of them, mini-preppred them with the new mini-prep kit, and recorded the resulting DNA concentrations. With these DNA tubes, he went ahead and made a transformation of the three plasmids into BL21 cells (which Alessandro had made throughout the past week). Vincent plated these transformed cells on kanamycin plates and incubated them overnight.
Since the previous day's Colony PCR had partly failed due to an extension time for lysis that was too short (1 minute rather than 2 minutes), Vincent made a Colony PCR for K1-T7-Lysis and a Colony PCR for C5-T7-RFP and C5-T7-Lysis. Perhaps because it was almost 9 in the evening, Vincent did not realize that he had successfully amplified both the K1 and the C5 T7 lysis, showing that the Gibson Assembly had in fact worked as expected. He saw the primer dimers at the bottom of the gel and did not think to even look for faint bands at the 2 kb level where the lysis piece would appear. So the Gibson did in fact work, but Vincent only found this out retrospectively on Tuesday of the following week (this wiki post is written with some tardiness).
Sunday, 21 August
Vincent took out the plates from the BL21 transformation. The results were spectacular: thousands of colonies. He then picked one colony from each plate to use for liquid cultures. He did not forget the antibiotic (as he usually does). The idea was to have liquid cultures ready for an IPTG platereader experiment on Monday.
Vincent also ran a miniprep on the C5-T7-RFP cultures, but got a very low yield of DNA (9-10 ng/uL).
Monday, 22 August 2011
Nadine made 2 Gibson reactions (equimolar ratios):
- LacI and pSB3K1 Pconst-TetR, to assemble pSB3K1 Pconst-TetR Ptet-LacI plasmid
- RFP and J61002 backbone, to assemble J61002 Plac-RFP plasmid
She also made negative controls, where only the backbone has been mixed with water, and transformed everything.
Vincent ran a platereader experiment using the liquid cultures of T7-RFP plasmids. Results should be appearing shortly.
Vincent received all 18 of the primers he needed to make the T7 promoter variants. Here is the list of these primers (this indexing will be used in the gel pictures):
1. T7 const
2. T7 014
3. T7 030
4. T7 054
5. T7 080
6. T7 111
7. T7 const (-7-12)
8. T7 const (-3-1/+1+3)
9. T7 const (-10)
10. T7 lac2
11. T7 lac2 014
12. T7 lac2 030
13. T7 lac2 054
14. T7 lac2 080
15. T7 lac2 111
16. T7 lac2 (-7-12)
17. T7 lac2 (-3-1/+1+3)
18. T7 lac2 (-10)
Vincent made 50 uM and 500 nM dilutions of the primers.
Tuesday, 23 August 2011
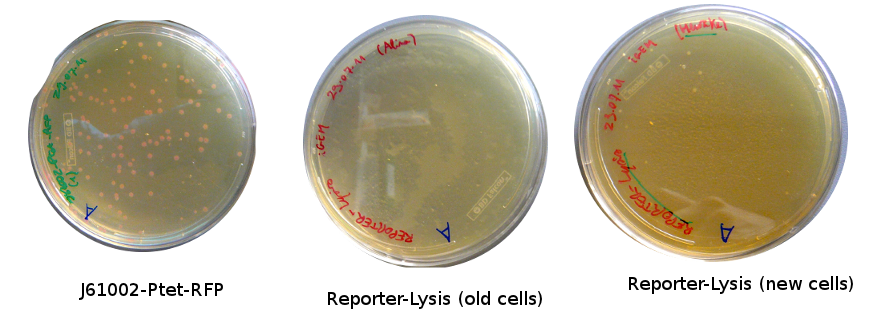
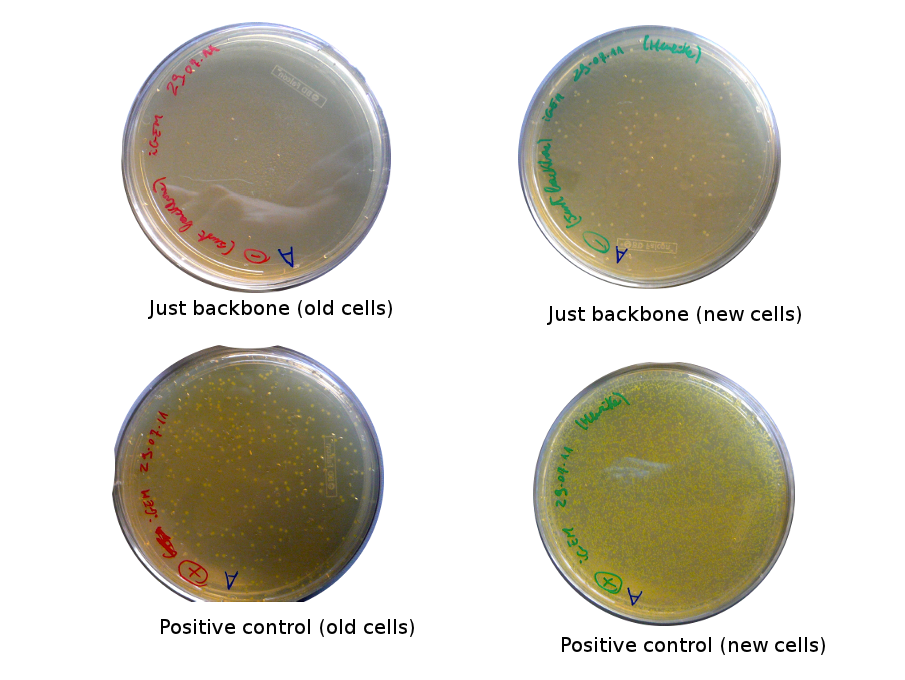
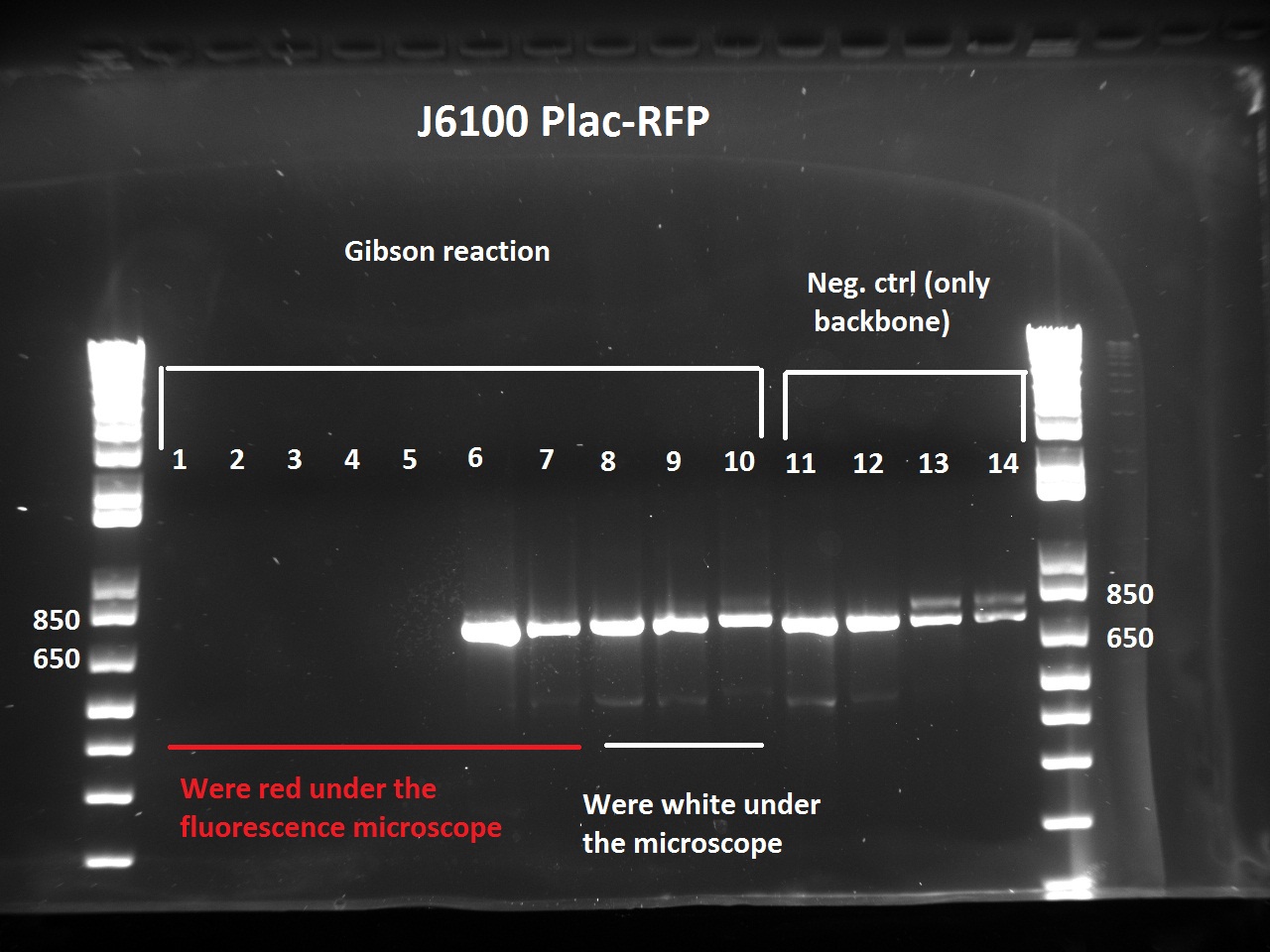
The Gibson-transformed cells yielded a lot of mutants, with significantly more on the Gibson plates than on the negative controls ones. One strange thing with the J61002 Plac-RFP: the negative control cells were weakly red fluorescent, while for the Gibson assembly some of the cells were expressing RFP. We'll see the results of the colony PCR tomorrow, but it's possible that the cells are autofluorescent...
Vincent made an extension PCR for both the RFP and Lysis T7 promoter variants (36 samples). The extension PCR had an elongation time of 1 min. or RFP and 2 min. for Lysis. The protocol is avaible in the protocol section.
Clearly the RFP T7 promoter variants went well, but the T7 Lysis promoter variants ... not so much. Vincent made liquid cultures of all the T7 lysis plasmids (both C5 and K1) and put them in the incubator. He also pelleted all the C5 RFP liquid cultures and the K1-Lys cultures that he had made on Monday (6 colonies per promoter) thinking that he had not yet achieved a full T7-lysis. The pellets are being kept in a special box called "Pellets".
Wednesday, 24 August 2011
Nadine ran the colony PCRs on a gel and got strange results.
- For J61002 Plac-RFP, the negative control cells are definitely red - although they should not contain RFP - and the colony PCR that includes a primer on RFP is positive. The colonies 8-10 were still white on the plate, but here again the colony PCR is positive. Perhaps this comes from the template plasmid used for the PCR (J61002 Ptet_EFP); since the miniprep failed we have to wait before sending the samples for sequencing.
- For pSB3K1 Pconst-TetR Ptet-LacI, we also have positive results for the negative control. Here it's unlikely that it can be because of the template plasmid for the PCR, because be used the repressilator that has ampicillin resistance when the plates contained kanamycin. We'll see tomorrow the results of the sequencing of colony 7.
Preparing for the next Gibson assemblies, Nadine also ran two PCRs:
- One for amplifying the lysis cassette and putting it into J61002 backbone (with a new primer)
- One for amplifying LacI and put it into J61002 Plac-RFP or Plac-lysis, once we'll have them.
Vincent mini-prepped the liquid cultures of the original K1-T7-Lys plasmids that had been (unknowingly) successful on Saturday. He then transformed these into BL21 cells and plated them. The C5-lys plasmid liquid cultures did not grow because Vincent had accidentally used gentamycin instead of chloramphenicol in his liquid cultures. He re-did the C5 cultures.
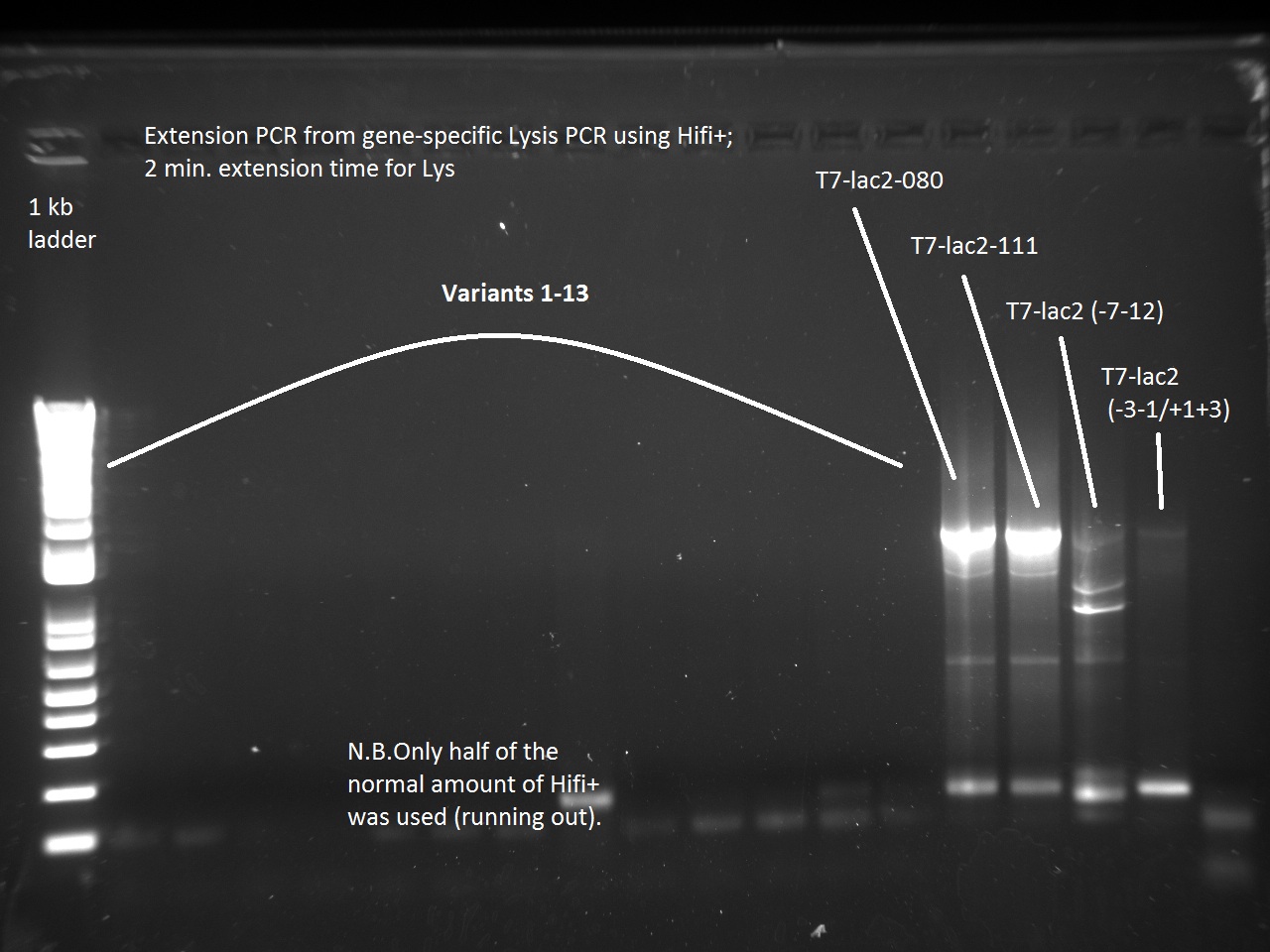
Vincent also ran a new set of 18 T7-Lys promoter variants using Extension PCR. Because we were running out of Hifi+ enzyme, we used only half of the enzyme requirement (9 uL instead of 18).
Apparently, 14 through 17 were amplified correctly.
Thursday, 25 August 2011
The sequencing results of the pSB3K1 TetR-LacI plasmid came. It seems that LacI got insterted correctly, but that TetR got lost... We'll try to send this plasmid again for sequencing with another primer to be sure. Also, a new Gibson assembly has been done. For the J61002 Plac-RFP plasmid, Nadine forgot the cell cultures on the bench so the sequencing will have to wait over the week-end. Here again, a new Gibson assembly has been done, along with J61002-lysis.
Vincent made liquid cultures from the successful transformation of the K1-T7-Lysis into BL21 cells. Nadine was kind enough to purify all 18 of Vincent's T7-RFP variants, as well as the 4 T7-Lysis variants. In the same spirit, the C5-T7-Lysis plasmids were miniprepped and then transformed into the BL21 cells and plated.
In order to continue with the production of T7 promoter variants, Vincent had to make more gene-specific Lysis (i.e. T4 Lysis with RBS upstream). For this, Vincent used the new Hifi+ enzyme that had been ordered the previous day. Since we were also running out of PCR primer mix, Vincent went ahead and made some more.
Friday, 26 August 2011
Vincent ran a plate-reader experiment for the K1-T7-lysis BL21 cells. The first three rows received various doses of IPTG before the experiment got under way. The next three rows (const, lac, lac2) received no IPTG for the first 2 hours of incubation but then were given the same IPTG doses as the first three rows. Data analysis will take place tomorrow.
Vincent also made liquid cultures of the C5-T7-lysis BL21 cells. After consultation with Henrike, we have decided to drop the C5 backbone since it does not really add to the work we are doing and only adds more PCRs and cultures.
Vincent made a Gibson assembly of the 18 T7-RFP promoter variants and the 3 T7-Lysis variants into the K1 backbone. The DNA will be transformed on Saturday.
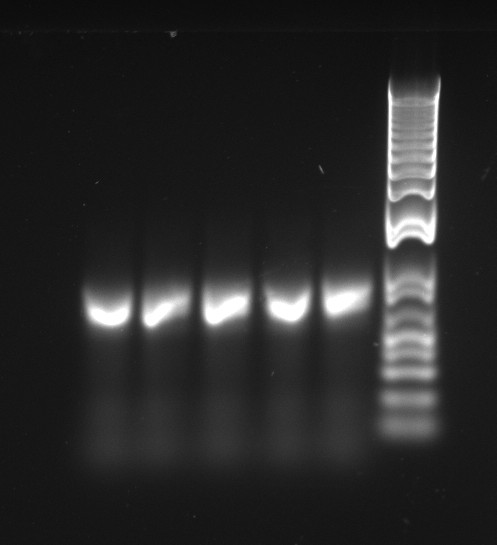
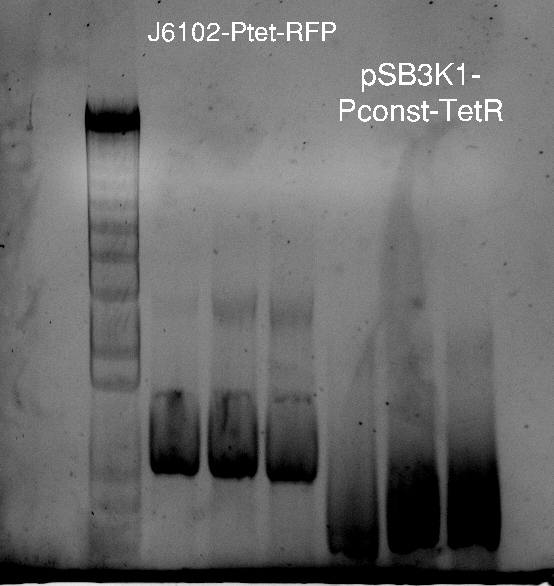
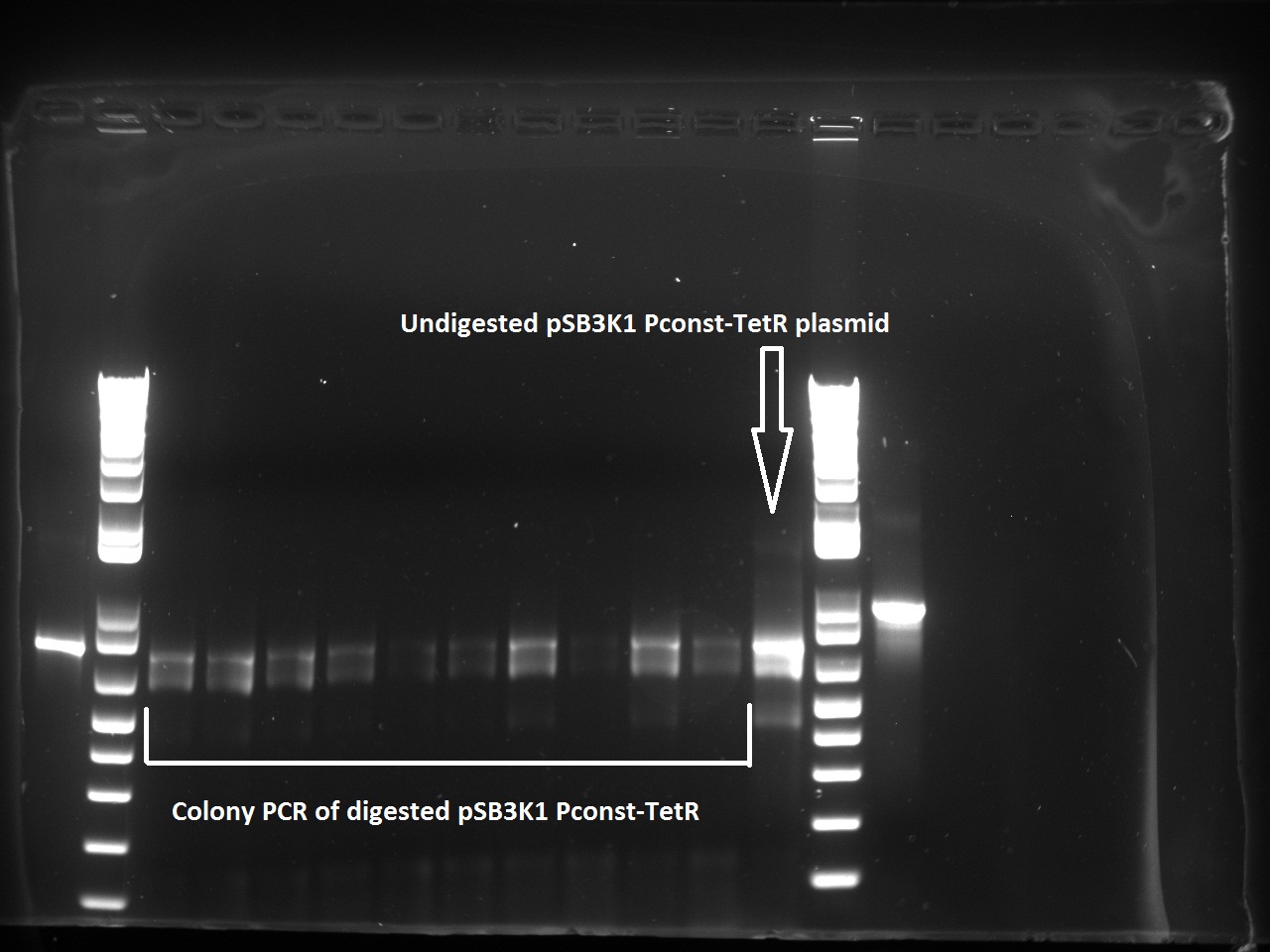
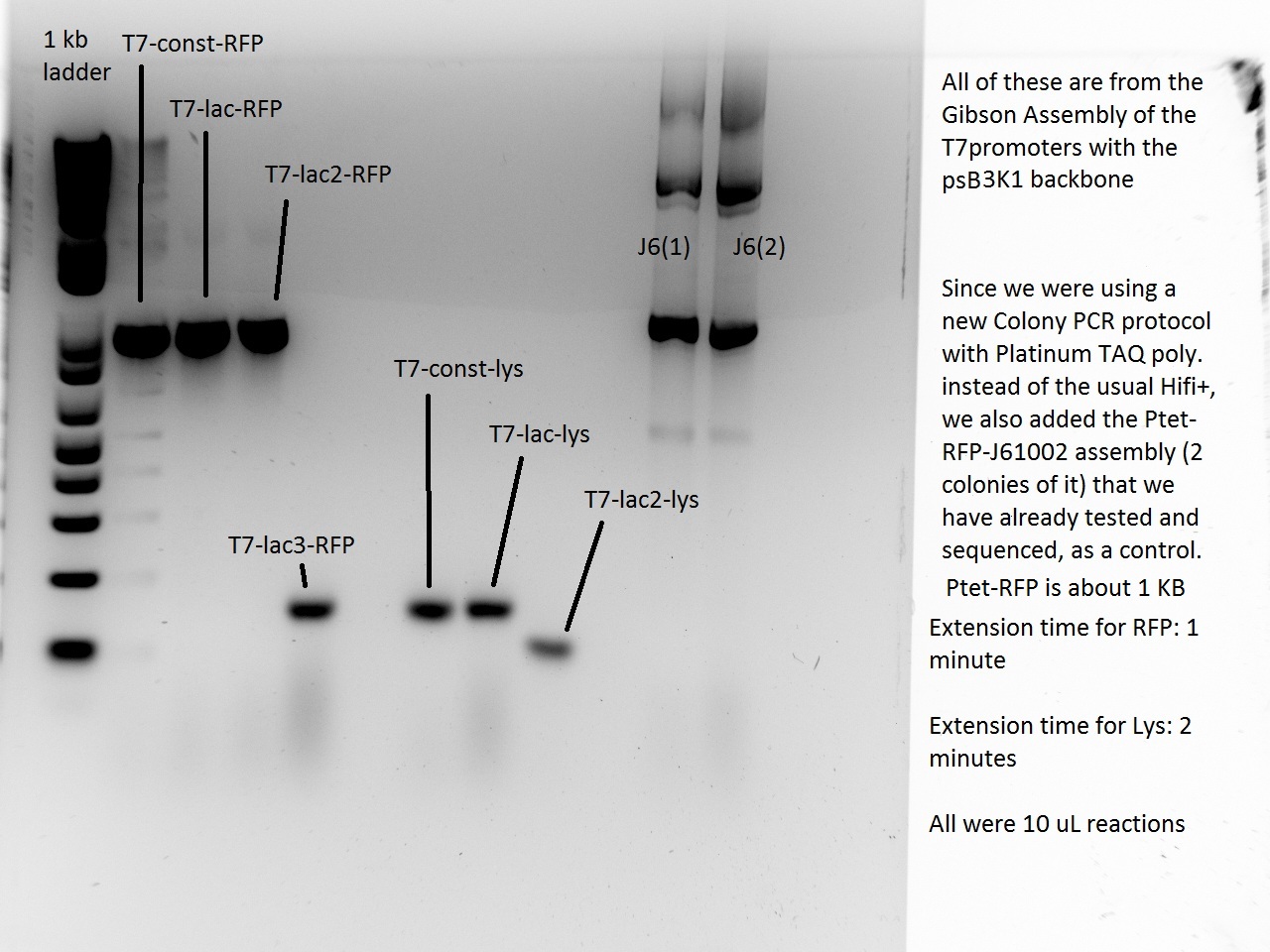
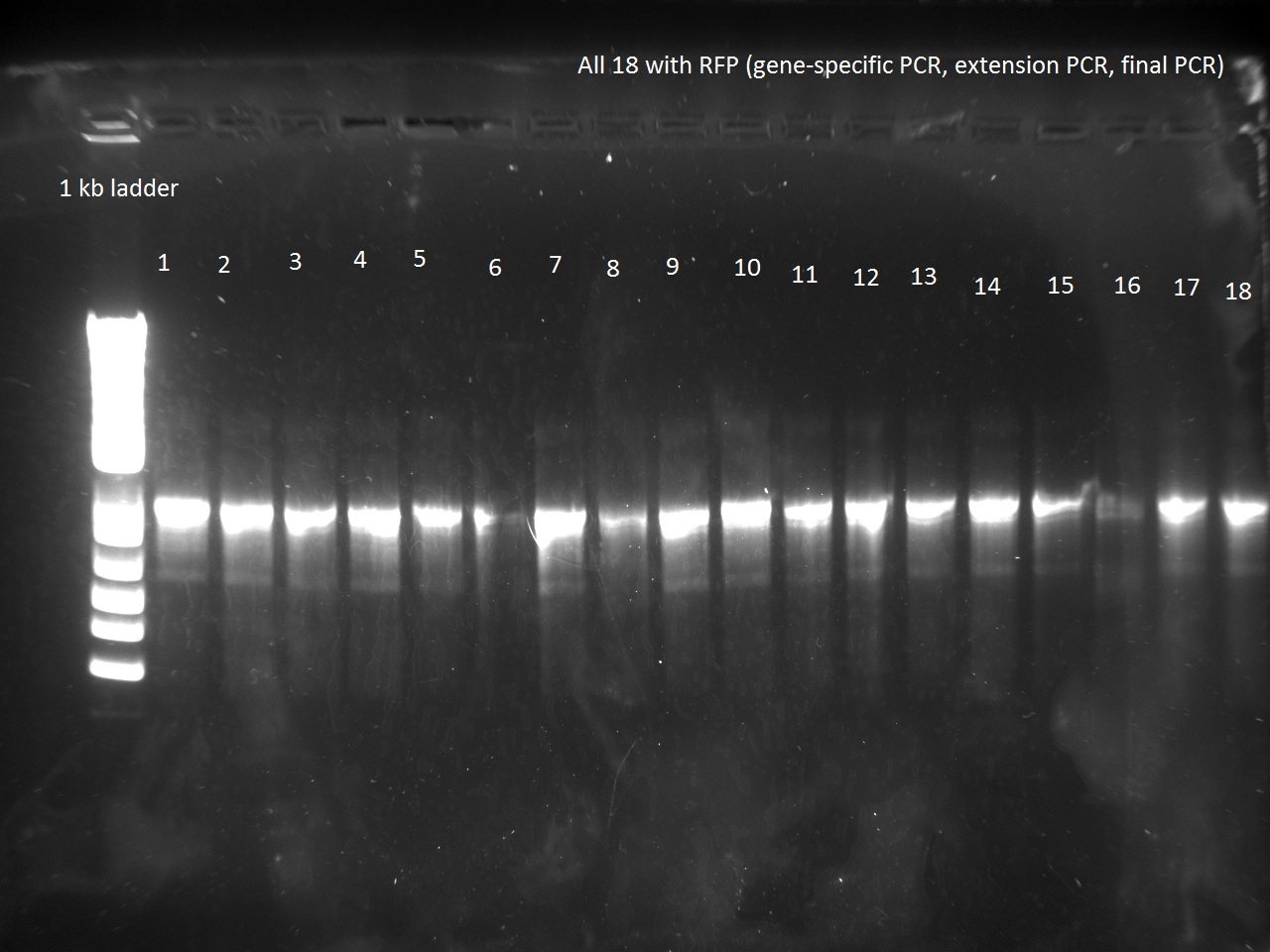
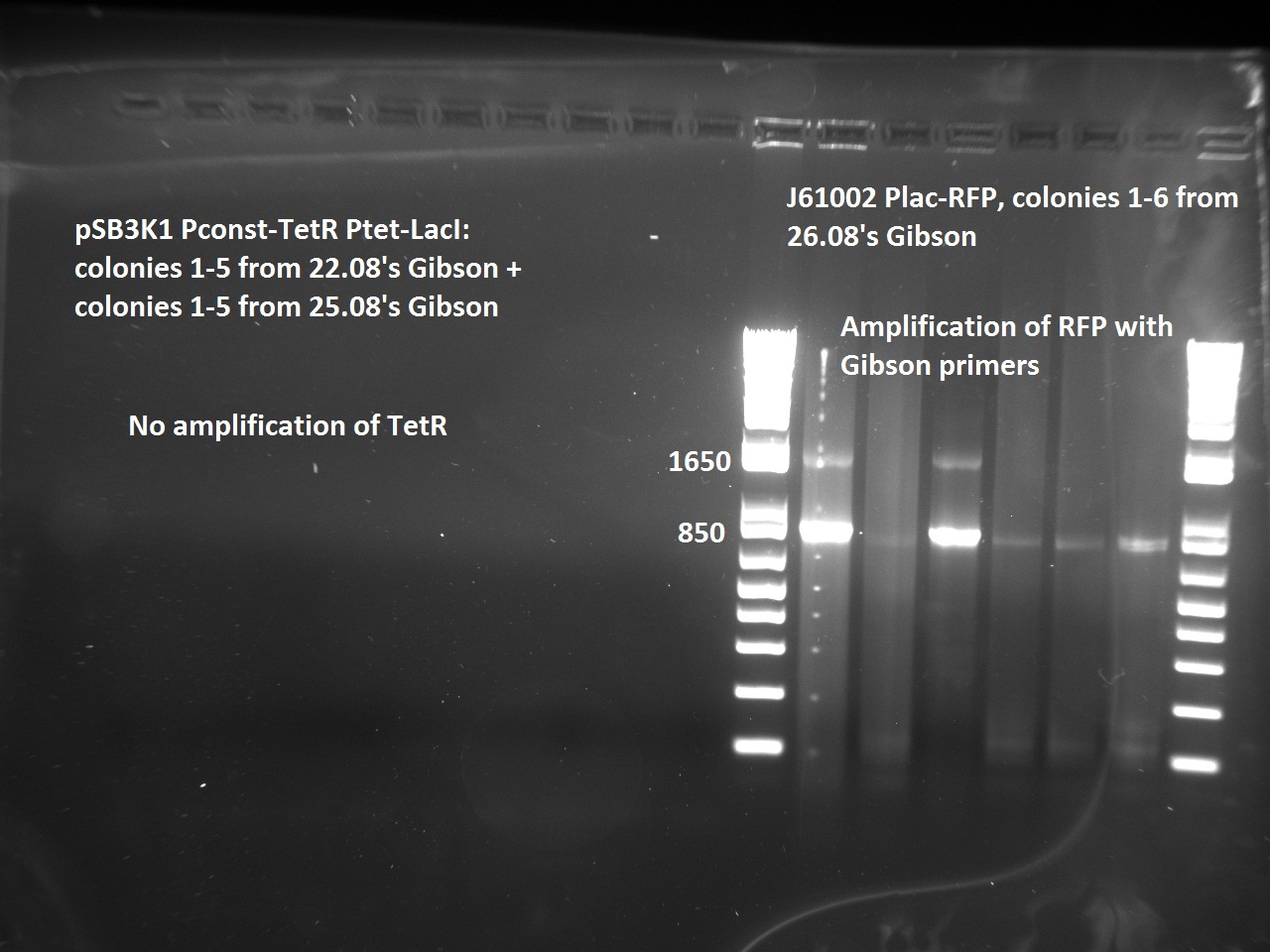
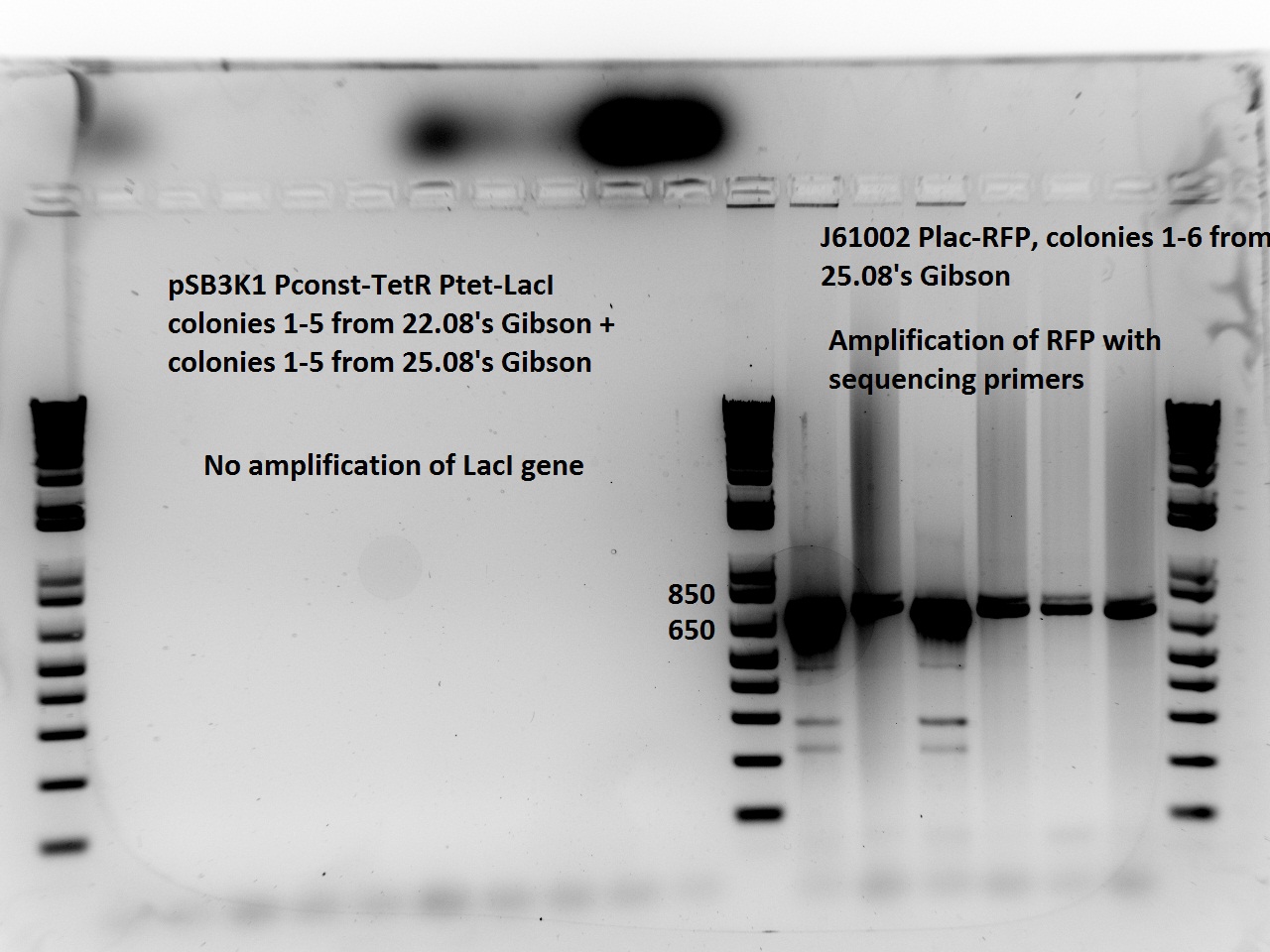
Nadine made colony PCRs for yesterday's Gibson assemblies: J61002 Plac-RFP and pSB3K1 TetR-LacI; she also made PCRs for pSB3K1 TetR-LacI that had been sent for sequencing on wednesday. Unfortunately, the plate with J61002 Plac-lysis yielded no colonies... Details of the PCRs:
- pSB3K1 TetR-LacI:
- amplify LacI with sequencing primers
- amplify TetR with sequencing primers
- J61002 Plac-RFP:
- amplify RFP with colony PCR primers
- amplify RFP with Gibson primers
With the sequencing primers, one of them binding in the middle of RFP, all 6 colony tested gave positive results. Strangely, there seems to be 2 bands with a slight difference in size. With the Gibson primers that contain Plac, colonies 1 and 3 have a brighter band. Either they are the only positive ones, or they just have a bigger amount of DNA (as also seen on the other gel).
For pSB3K1, both PCRs failed. Since it's very unlikely that we have a plasmid that lost TetR, there must have been a problem in the PCR; it will be repeated.
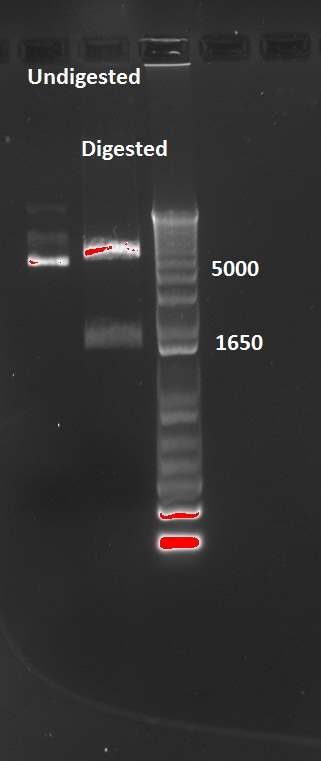
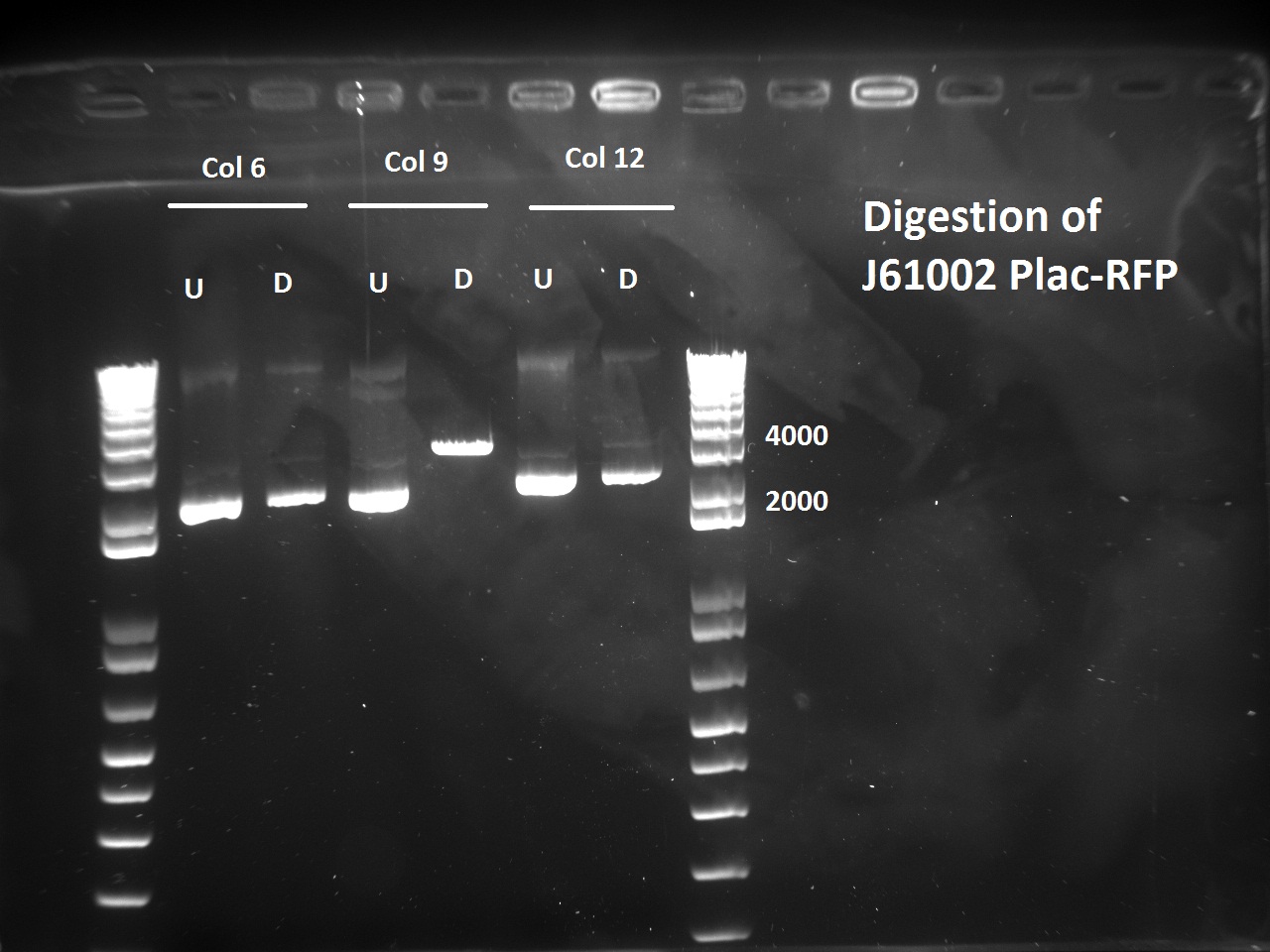
Nadine also digested the (supposed) J61002 Plac-RFP plasmids from Tuesday's Gibson assembly: colony 6 from Gibson plate that was red, colony 9 from Gibson plate that was white and colony 12 from negative control plate that was also red. Pst1 has a restriction site in the original J61002 backbone, therefore it should cut independently of RFP insertion. The results indicate that only colony 9 was digested by the enzyme, therefore it's likely that the red colonies contain an unknown plasmid that has RFP... Since colony 9 was positive for RFP colony PCR, it can be our expected plasimid that has little transcription of RFP.
 "
"