Team:Cornell/Action
From 2011.igem.org
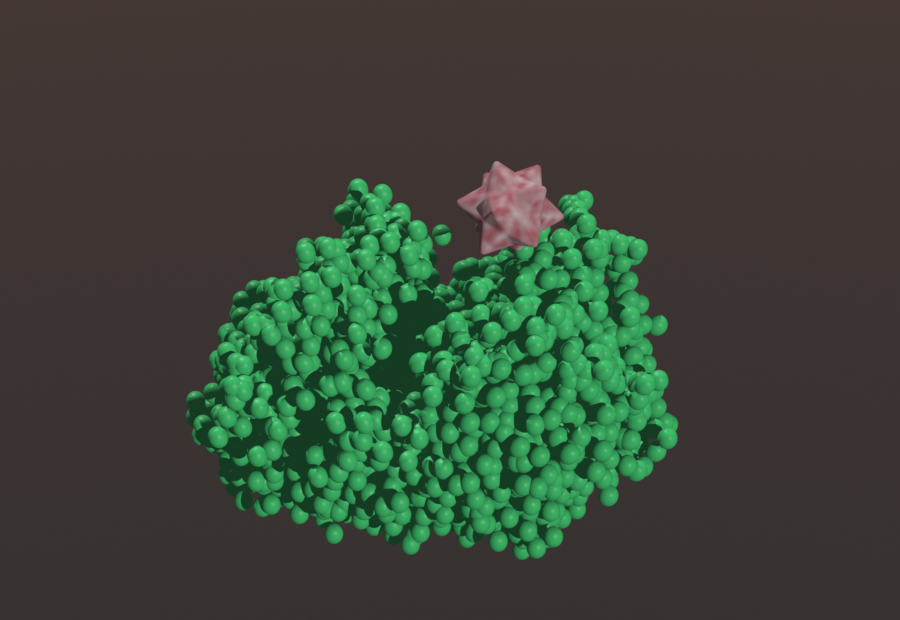
Original Description:
- Substrate represented by different geometric shapes flow through enzyme pathway
- As substrate flows through each stage, it is processed by the corresponding enzyme
- Substrate starts as a sphere, becomes an octagon, becomes a dodecahedron, and ends as a stellated icosahedron.
Interpreted Components:
1. Proteins
i. VioA , VioB, VioE – attractive, low resolution
2. Substrates
i. Easily distinguishable shapes.
ii. Organic textures
3. Protein-‐substrate interaction animation
i. Morphmaps that look like the enzymes are ‘nomming’ the substrate.
ii. Proper timing for substrate entering enzyme bonding site and start of processing
4. Side by side animations
i. Learn Final Cut Pro
Components Development Summary
1. Proteins
- Original attempts at creating proteins used mMaya to create point clouds from xyz data found in PDB databases online. Simple meshes were replicated into the point cloud to create convincing proteins.
- File formatting prohibited the use of these proteins in our transition to modo. Modo also lacks a streamlined method to produce proteins. Subdivided meshes were layered onto the Maya point clouds to create proteins usable in modo, but the results were lacking aesthetically.
- Finetuning variables allowed creation of smooth and and high-‐resolution proteins that usable in modo. These were also able to be ported into other scenes.
2. Attractive yet low-‐resolution proteins
- Standardization of the proteins was across our scenes to make the animations feel integrated. Some scenes used many proteins and yielded geometric caches that were unwieldy to work with. The need for creating low-‐resolution proteins was thus born.
- Python scripts paired with the software Blender created reasonably realistic proteins from PDB files that still had low polygon counts.
3. Substrates – Easily identifiable shapes
i. Creating the platonic solids using modeling software was redolent of geometry classes and took a surprising amount of time to make look both attractive and recognizable. Classical geometric construction techniques were used to make each of the desired polyhedrons
4. Substrate textures
i. The textures for the solids were created with a diffusion layer set to noise that produces a gradient of magenta colors. Adjustment of the frequency parameters of noise, the layer locator, and color parameters yielded an organic looking texture .
5. Protein-‐substrate interaction animation
i. As it was necessary for modeled proteins to vary from an ‘open’ to ‘closed’ position (see animation), morph deformers became an essential tool. Keyframing the opacity of the morphmap would change severity with which the morph is applied. This creates a very smooth and controllable transition between the two modeled states.
6. Realistic morphmaps
i. Original attempts at ‘closed’ protein states looked very rigid and mechanical. This is because the components in each group selected moved by a similar amount, a very naïve approach
ii. Future attempts utilized falloffs to make the opening and closing of the protein look organic and elastic.
iii. Getting realistic timing for the substrate movement and toggling of the morphmap was simply an exercise in patience and persistence.
7. Creating side by side protein animation
i. Each protein was individually animated processing its respective substrate. This made the animations of each protein loopable and easier to animate/render.
ii. Final Cut Pro was learned and utilized to edit the three final video clips to combine them into a single clip
Summary of Scene Progression
This scene was done last as it depended on the final proteins being used for the rest of the animations. Much of the work involved modeling proteins.
1. Protein modeling. (06/27/2011 -‐ 08/10/2011)
Created mMaya versions of VioA, VioB, VioE, and streptavidin
Highly triangulated ports of these proteins were made to modo
High quality proteins were made for modo
Reduced polygon counts using Maya tools reduced geometric memory use in modo by half
Blender scripts utilized to create even lower polygon count proteins from pdb files
3. 1st draft – Crude hinge morph maps (08/14/2011 – 08/15/2011)
Creating very primitive morphmaps, an animation using the same protein colored three different colors
was made to illustrate the concepts that would need to be utilized for the final scene. Keyframes were
eyeballed and positions were manually placed.
Despite rudimentary techniques, reception from the team was highly positive.
3. 2nd draft – Replicated proteins (08/17/2011 – 08/20/2011)
Pleased with initial success, proteins were replicated and given time delays to create an entire
screen of proteins processing substrate.
Reception was still positive, but this didn’t have the same visual appeal as the initial single protein
animation. It was advised to go back to a simple single protein model.
4. 3rd Draft – Full pathway, three proteins. (07/11/2011 -‐ 07/15/2011)
Scene was rebuilt using the feedback from above. The single protein perstage simplified the appearance of the animation, making more clear what was occurring. The morphmaps were created with move and falloff commands until they looked reasonable. Each of the three stages was animated individually and combined using Final Cut Pro.
Reception and Review: Positive. The current animation illustrates the process well, and is visually pleasing.
Future versions may include a remake using our very high resolution proteins.
 "
"