Team:Caltech/Week 7
From 2011.igem.org
| (12 intermediate revisions not shown) | |||
| Line 94: | Line 94: | ||
<td>23.4</td> | <td>23.4</td> | ||
</tr> | </tr> | ||
| - | |||
</table> | </table> | ||
The gel gave no bands in any lane, with or without the restriction digest. pNT003 + without the Spe1 digest had a faint smear, the other lanes had nothing. Chose to transform anyway<br/> | The gel gave no bands in any lane, with or without the restriction digest. pNT003 + without the Spe1 digest had a faint smear, the other lanes had nothing. Chose to transform anyway<br/> | ||
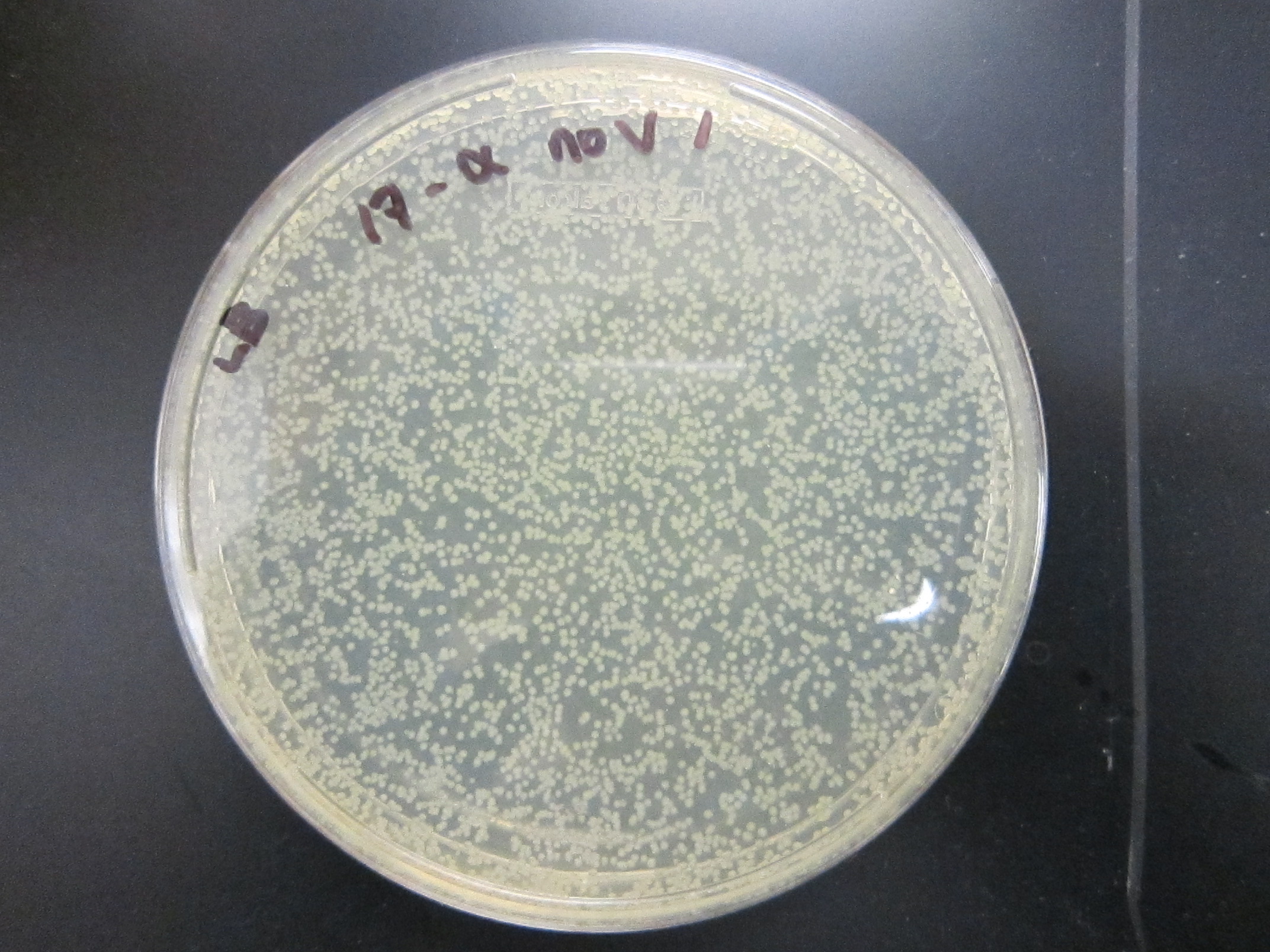
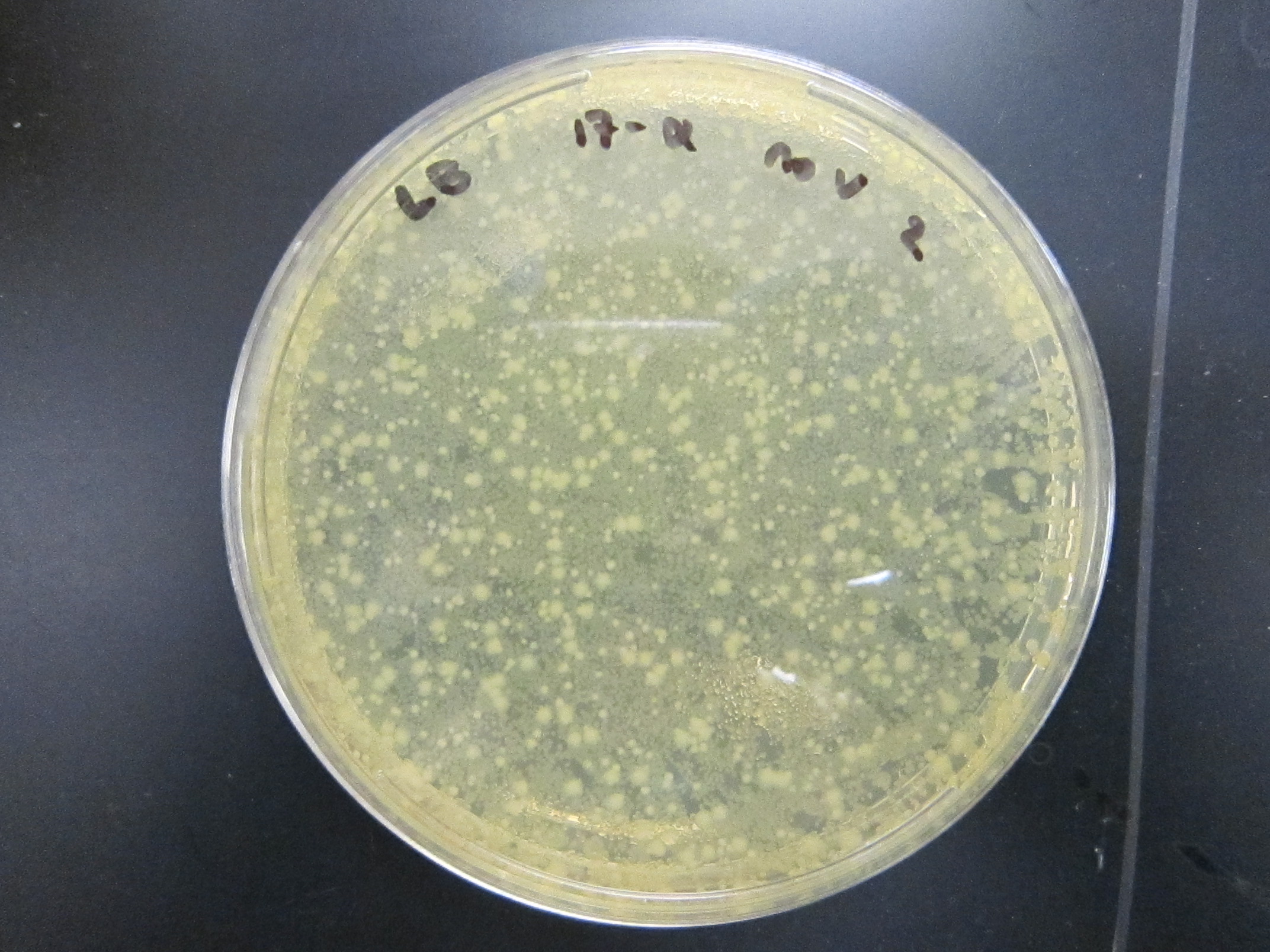
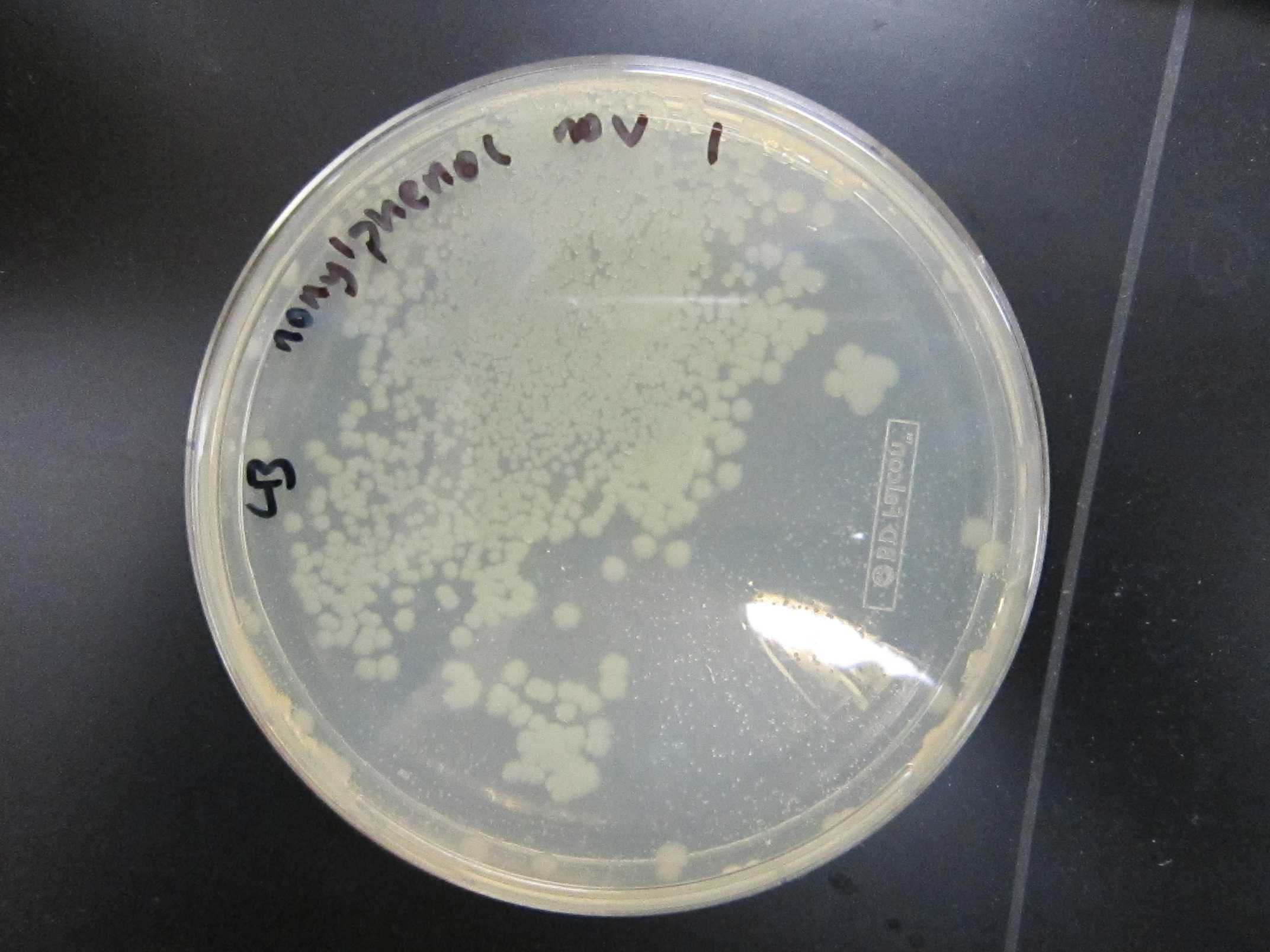
| - | + | All enrichment cultures but one showed growth on LB.<br/> | |
| - | All enrichment cultures but one showed growth on LB. | + | |
| - | + | ||
<gallery> | <gallery> | ||
File:17a_+V_1.jpg|17a-ethinylestradiol +V | File:17a_+V_1.jpg|17a-ethinylestradiol +V | ||
| Line 138: | Line 135: | ||
Continued attempts to spread chemical solution on minimal media plates<br/> | Continued attempts to spread chemical solution on minimal media plates<br/> | ||
Minimal media transfers <br/> | Minimal media transfers <br/> | ||
| + | |||
===Results=== | ===Results=== | ||
| + | |||
<gallery> | <gallery> | ||
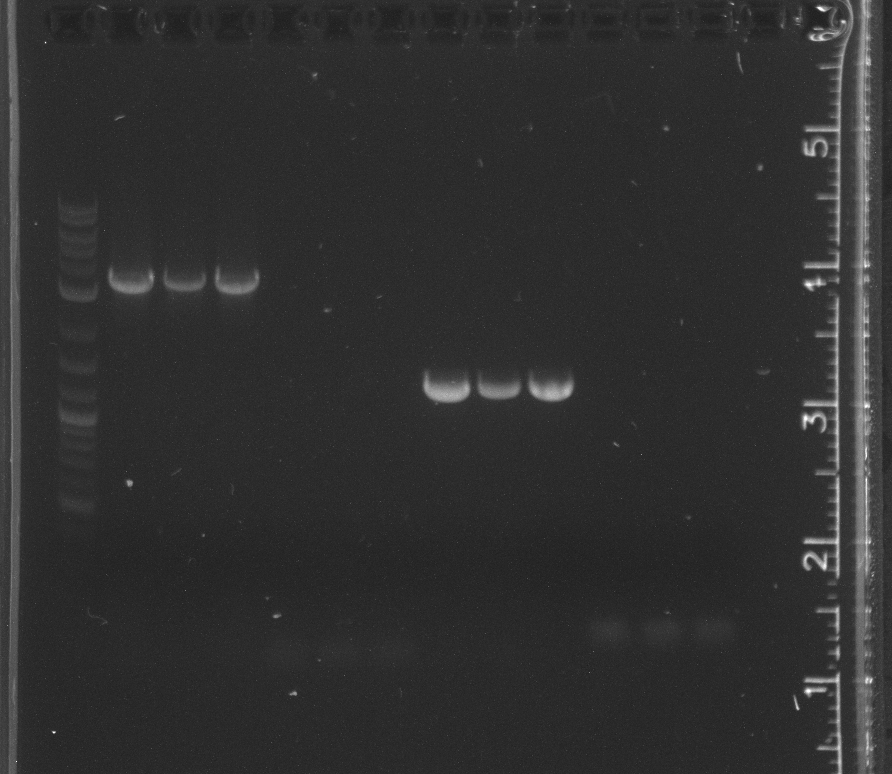
File:7-2716sgel.jpg|lane 1 NEB 2-log ladder, 2-3 negative controls, 4-6 LA River sample 9, 7-9 LA River sample 10, 11-12 pET53DEST | File:7-2716sgel.jpg|lane 1 NEB 2-log ladder, 2-3 negative controls, 4-6 LA River sample 9, 7-9 LA River sample 10, 11-12 pET53DEST | ||
File:7-27psb3k3.jpg|lane 1 NEB 2-log ladder, 2-4 pSB3K3 | File:7-27psb3k3.jpg|lane 1 NEB 2-log ladder, 2-4 pSB3K3 | ||
</gallery> | </gallery> | ||
| + | |||
The 16s PCR from yesterday, shown in the gel today, had a band in the first lane of negative controls, indicating contamination.<br/> | The 16s PCR from yesterday, shown in the gel today, had a band in the first lane of negative controls, indicating contamination.<br/> | ||
| + | |||
The pSB3K3 amplification failed. The bands indicated a much lower length than expected. After doing an alignment in Geneious, it appears our pSB3N5 primers we ordered for pSB3C5, which we are no longer using because of our competent cell strain, do not work with earlier versions of the 3 origin plasmids.<br/> | The pSB3K3 amplification failed. The bands indicated a much lower length than expected. After doing an alignment in Geneious, it appears our pSB3N5 primers we ordered for pSB3C5, which we are no longer using because of our competent cell strain, do not work with earlier versions of the 3 origin plasmids.<br/> | ||
| + | |||
Both the negative control and experimental of PCR purified Gisbson assembly from yesterday of pNT003 had 0 colonies. The transformation with 1 ul pUC 19 had no colonies.<br/> | Both the negative control and experimental of PCR purified Gisbson assembly from yesterday of pNT003 had 0 colonies. The transformation with 1 ul pUC 19 had no colonies.<br/> | ||
| + | |||
<table border="1"> | <table border="1"> | ||
<tr> | <tr> | ||
| Line 162: | Line 165: | ||
BPA solutions fail to form uniform layers when poured on top of agar, but form a uniform layer when poured directly onto a clean plate. | BPA solutions fail to form uniform layers when poured on top of agar, but form a uniform layer when poured directly onto a clean plate. | ||
| + | |||
| + | <gallery> | ||
| + | File:BPA_on_agar.jpg|BPA on agar | ||
| + | File:BPA_clean_plate.jpg|BPA on clean plate | ||
| + | </gallery> | ||
| + | |||
==July 28== | ==July 28== | ||
| - | Plate packaged fosmids to obtain titer <br/> | + | Plate packaged fosmids to obtain titer<br/> |
| - | Set up enzyme binding assay of p450s <br/> | + | Set up enzyme binding assay of p450s with 17-α estradiol, bispehnol A, and nonylphenol with cytochrome p450-BM3 enzymes WT-F87A, H2A10, and 9-10ATSF87A<br/> |
Try out plating chemicals on bottom of plates with agar bacteria suspension on top <br/> | Try out plating chemicals on bottom of plates with agar bacteria suspension on top <br/> | ||
Miniprepped pSB4A5 from overnight culture of glycerol stock-54 ng/ul<br/> | Miniprepped pSB4A5 from overnight culture of glycerol stock-54 ng/ul<br/> | ||
Set up restriction digests of pNT002 insert and pSB3K3-1 with EcoRI-HF and PstI according to NEB protocol <br/> | Set up restriction digests of pNT002 insert and pSB3K3-1 with EcoRI-HF and PstI according to NEB protocol <br/> | ||
| + | Ligated pNT002 and pSB3K3 following NEB protocol <br/> | ||
Find positive control for transformation, test transformation<br/> | Find positive control for transformation, test transformation<br/> | ||
Run gel of 16s and vector. Dpn1 digest vector.<br/> | Run gel of 16s and vector. Dpn1 digest vector.<br/> | ||
| Line 173: | Line 183: | ||
===Results=== | ===Results=== | ||
<p>Yesterday's 16s PCR failed, as there were no bands in control or experimental lanes. The vector was amplified and was the correct length. We redid it, but again there were no bands in the control or experimental lanes.</p> | <p>Yesterday's 16s PCR failed, as there were no bands in control or experimental lanes. The vector was amplified and was the correct length. We redid it, but again there were no bands in the control or experimental lanes.</p> | ||
| + | Enrichment cultures plated in minimal media with chemical layer, to be checked in a few days.<br/> | ||
Gel extraction of pNT003 insert was not much better than yesterday. Here are the concentrations:<br/> | Gel extraction of pNT003 insert was not much better than yesterday. Here are the concentrations:<br/> | ||
<table border="1"> | <table border="1"> | ||
| Line 188: | Line 199: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | ==July 29== | ||
| + | Attempted to PCR pNT002 and pNT003 inserts from the purified gel extractions of July 27, using the same primers.<br/> | ||
| + | Attempted to PCR pNT003 insert from component parts with a gradient of DMSO to prevent non-specific priming and hopefully amplify the correct length insert instead of the 400bp band that keeps appearing.<br/> | ||
| + | Titering of packaged fosmid inserts at concentrations 1x, .1x, .01x, and .001x on LB-chlor plates<br/> | ||
| + | Analysis of p450 assay with reverse-phase HPLC<br/> | ||
| + | Tried longer ligation time for pNT002 insert/pSB3K3<br/> | ||
| + | ===Results=== | ||
| + | HPLC reveals shift in the chemical structure of BPA after incubation with p450's WT-F87A and 9-10ATSF87A from a peak of 5.26 to 6.5<br/> | ||
| + | <gallery> | ||
| + | File:Blank_BPA.jpg|HPLC UV peak for BPA dissolved in DMSO | ||
| + | File:WT-F87A.jpg|HPLC UV peak for BPA degraded by WT-F87A (a p450 enzyme) | ||
| + | File:9-10ATSF87A.jpg|HPLC UV peak for BPA degraded by 9-10ATSF87A (a p450 enzyme) | ||
| + | </gallery> | ||
| + | No colonies on pNT002/pSB3K3 ligation <br/> | ||
| + | ==July 30== | ||
| + | Take out plates from fosmid titering<br/> | ||
| + | Ran gels of PCR of pNT002 insert, pNT003 insert and pSB3K3 from yesterday.<br/> | ||
| + | Mass spectroscopy of bisphenol A's p450 degradation products | ||
| + | ===Results=== | ||
| + | <p>Two colonies on both 9mix and 10-2mix fosmid 1x dilutions</p> | ||
| + | <gallery> | ||
| + | File:7-30pcrgel1.jpg|lane 1 NEB 2-log ladder, 2-4 pNT002 insert, 5-7 pNT003 insert, 8-10 pSB3K3 | ||
| + | File:7-30pcrgel2.jpg|PCR of pNT003 insert with increasing DMSO:lane 1 NEB 2-log ladder, 2-3 0% DMSO, 4-5 0.5%, 6-7 1%, 8-9 2%, 10-11 3% | ||
| + | </gallery> | ||
| + | pSB3K3 was not amplified. The DMSO does not to appear to have decreased the 400 bp band or increased the correct 2 kb band.<br/> | ||
| + | No colonies from pNT002/pSB3K3 ligation.<br/> | ||
| + | BPA samples did not ionize well in LCMS; retry with GCMS.<br/> | ||
}} | }} | ||
Latest revision as of 20:58, 18 August 2011
|
Project |
July 24Started overnight cultures of pSB3K3 July 25PCR'd parts for pNT002: R0040, K123001, B0014, pSB4A5 ResultsDecided not to use the pNT003 PCR'd insert, as the band of the correct length was much fainter than a lower length band. Chose not to do Gibson assembly of the positive GFP control, despite not having any green colonies last week, instead focusing on assembling pNT002 and pNT003. PCR concentrations
PSB3K3 miniprep concentrations
July 26Packaging of 9mix and (10mix + 10-2) ligations Results
The gel gave no bands in any lane, with or without the restriction digest. pNT003 + without the Spe1 digest had a faint smear, the other lanes had nothing. Chose to transform anyway
July 27Grew up cells for titering of packaged fosmids; reached OD600 of 0.974 ResultsThe 16s PCR from yesterday, shown in the gel today, had a band in the first lane of negative controls, indicating contamination. The pSB3K3 amplification failed. The bands indicated a much lower length than expected. After doing an alignment in Geneious, it appears our pSB3N5 primers we ordered for pSB3C5, which we are no longer using because of our competent cell strain, do not work with earlier versions of the 3 origin plasmids. Both the negative control and experimental of PCR purified Gisbson assembly from yesterday of pNT003 had 0 colonies. The transformation with 1 ul pUC 19 had no colonies.
BPA solutions fail to form uniform layers when poured on top of agar, but form a uniform layer when poured directly onto a clean plate. July 28Plate packaged fosmids to obtain titer ResultsYesterday's 16s PCR failed, as there were no bands in control or experimental lanes. The vector was amplified and was the correct length. We redid it, but again there were no bands in the control or experimental lanes. Enrichment cultures plated in minimal media with chemical layer, to be checked in a few days.
July 29Attempted to PCR pNT002 and pNT003 inserts from the purified gel extractions of July 27, using the same primers. ResultsHPLC reveals shift in the chemical structure of BPA after incubation with p450's WT-F87A and 9-10ATSF87A from a peak of 5.26 to 6.5
No colonies on pNT002/pSB3K3 ligation July 30Take out plates from fosmid titering ResultsTwo colonies on both 9mix and 10-2mix fosmid 1x dilutions pSB3K3 was not amplified. The DMSO does not to appear to have decreased the 400 bp band or increased the correct 2 kb band.
|
 "
"