Team:Caltech/Week 4
From 2011.igem.org
(Created page with "{{Team:Caltech/templateheader| Content= __NOTOC__ ==This Week== Get the BioBricks we ordered from the parts registry ([http://partsregistry.org/Part:BBA_K364309 K364309], [http:/...") |
|||
| (15 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
Content= | Content= | ||
__NOTOC__ | __NOTOC__ | ||
| - | == | + | ==July 3== |
| - | + | <p>Checked Gibson transformations<br/> | |
| - | + | Plated remaining transformed cells<br/> | |
| - | Continue | + | Made new dilutions of original enrichment cultures (BPA, nonylphenol, 17-alpha-ethinyl-estradiol, DDT)</p> |
| - | + | ||
| - | + | ===Results=== | |
| - | + | There was no growth on the transformed Gibson plates. Since we did a small reaction, we used all of the reaction in transforming competent cells. The Gibson assembly likely failed, as one of the negative control plates had a single colony. However, we have had trouble with our competent cells in the past and will try plating what we have left.<br/><br/> | |
| + | Gibson Transformations | ||
| + | <table border="1"> | ||
| + | <tr> | ||
| + | <th>Plate</th> | ||
| + | <th>Number of Colonies</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT001 +</td> | ||
| + | <td>0</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT001 -</td> | ||
| + | <td>0</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT002 +</td> | ||
| + | <td>0</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT002 -</td> | ||
| + | <td>1</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | ==July 4== | ||
| + | ===Results=== | ||
| + | After plating the remaining 450 ul of our transformed cells: | ||
| + | <table border="1"> | ||
| + | <tr> | ||
| + | <th>Plate</th> | ||
| + | <th>Number of Colonies</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT001 +</td> | ||
| + | <td>10</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT001 -</td> | ||
| + | <td>0</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT002 +</td> | ||
| + | <td>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT002 -</td> | ||
| + | <td>0</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | ==July 5== | ||
| + | <p>Start overnight cultures of Gibson transformations<br/> | ||
| + | Start gel to figure out length of purified river DNA (for fosmid kit)<br/> | ||
| + | Confirm we have enough DNA for work with estrogen receptor<br/> | ||
| + | Streak out new biobricks on chloramphenicol plates</p> | ||
| + | |||
| + | ===Results=== | ||
| + | The gel of the river DNA was inconclusive, as no bands were visible (even the weight ladder). The gel will be rerun. | ||
| + | |||
| + | ==July 6== | ||
| + | <p>Miniprep Gibson transformations and send off for sequencing<br/> | ||
| + | Redo gel for fosmid kit<br/> | ||
| + | Continue sequencing of estrogen receptor (using new forward primer and old reverse primer)<br/> | ||
| + | Start overnight cultures of new biobricks<br/> | ||
| + | Plate enrichment cultures on LB | ||
| + | </p> | ||
| + | |||
| + | ===Results=== | ||
| + | Again, no bands were visible on the gel. We will run the samples on a smaller gel to check the size of the DNA and confirm that there is in fact DNA in the sample. | ||
| + | |||
| + | ==July 7== | ||
| + | <p>Miniprep new biobricks and send off for sequencing<br/> | ||
| + | Meet with Pedro about HPLC and p450s<br/> | ||
| + | Image enrichment culture plates<br/> | ||
| + | Image second fosmid kit gel<br/> | ||
| + | Run river DNA on small gel to check size<br/> | ||
| + | Start new minimal media cultures from plates<br/> | ||
| + | Design primers for estrogen receptor test construct<br/> | ||
| + | |||
| + | ===Results=== | ||
| + | [[Image:River_gel_small.jpg|thumb|River samples run on a small gel. 1: ladder; 2:blank; 3: sample 9; 4: sample 10 ]] | ||
| + | The smaller gel shows a smear at the top of the gel for one of the samples, indicating that this sample contains DNA that is probably of the correct size for work with the fosmid kit.<br/> | ||
| + | |||
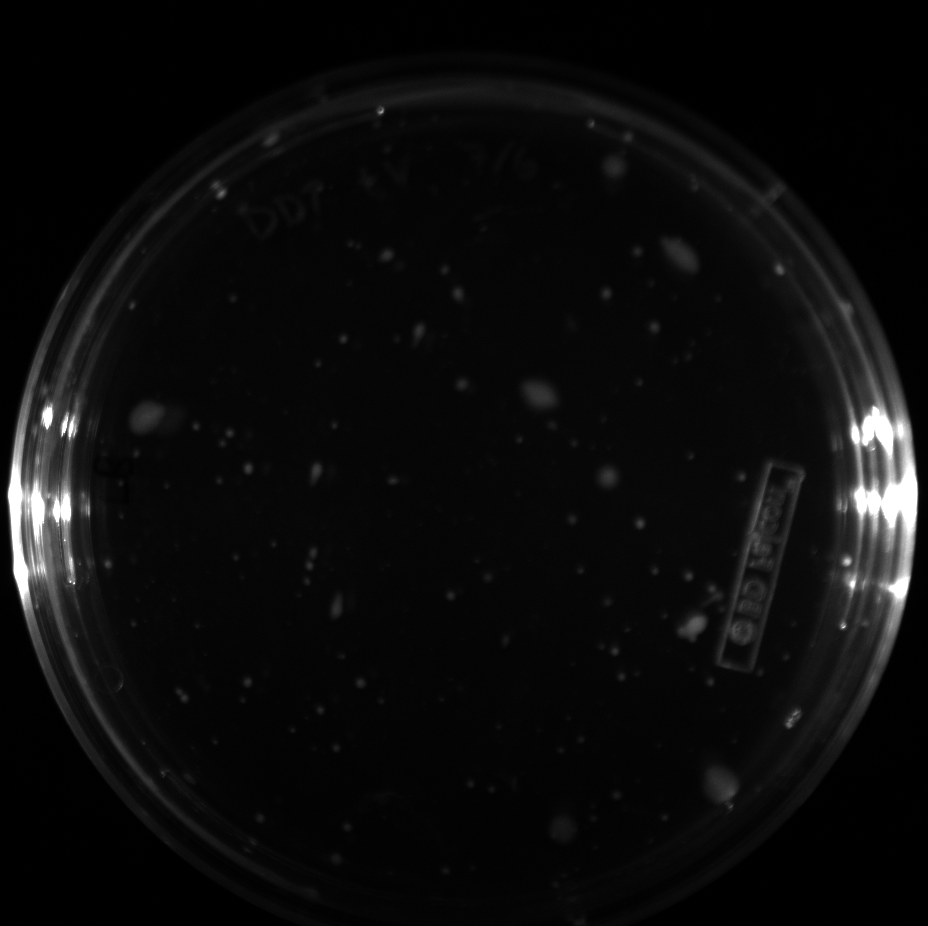
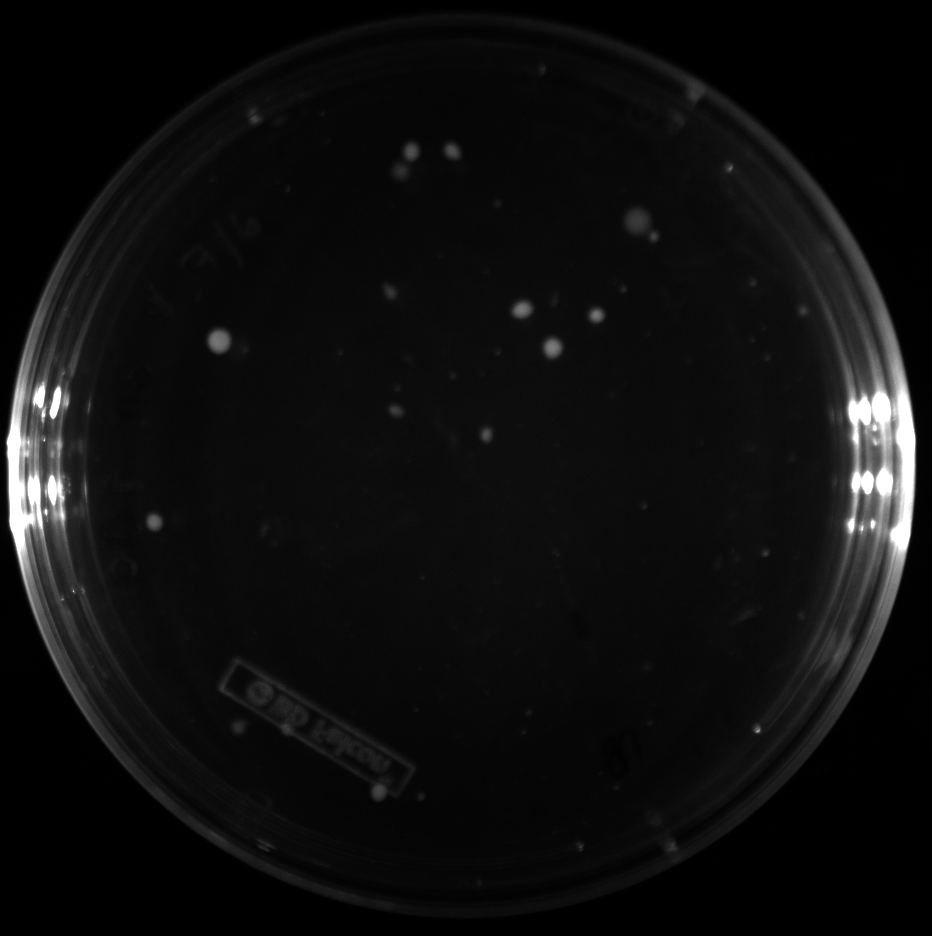
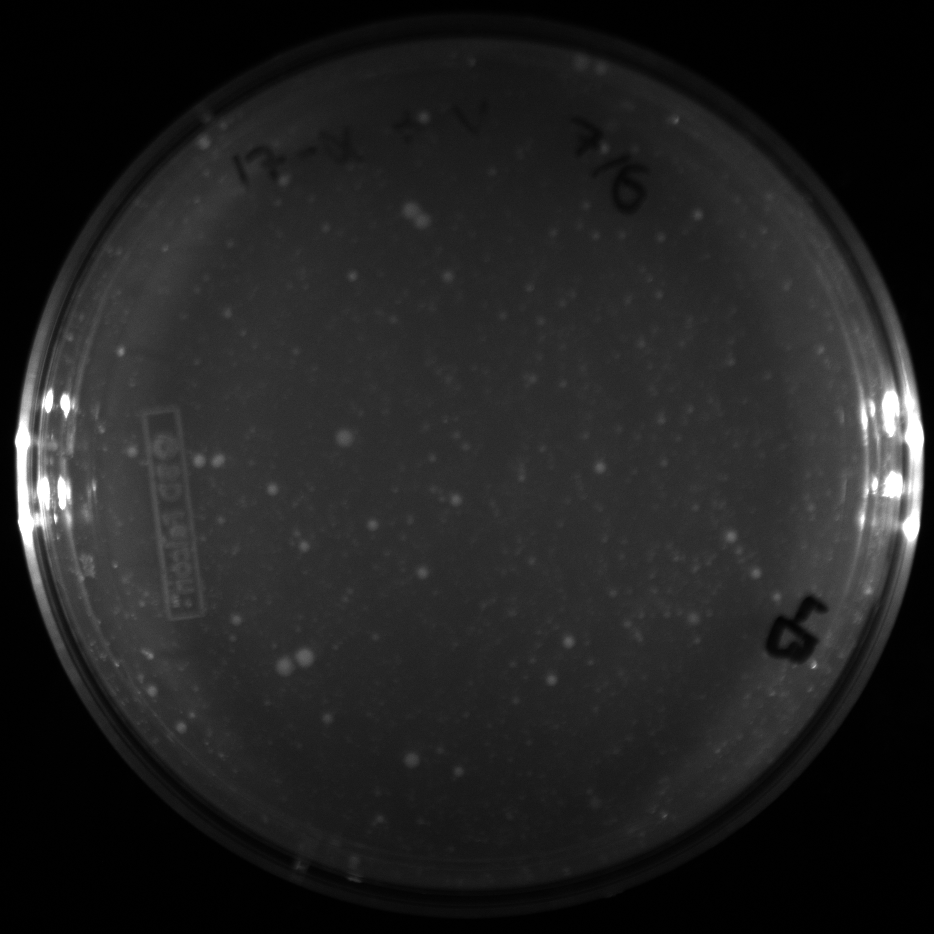
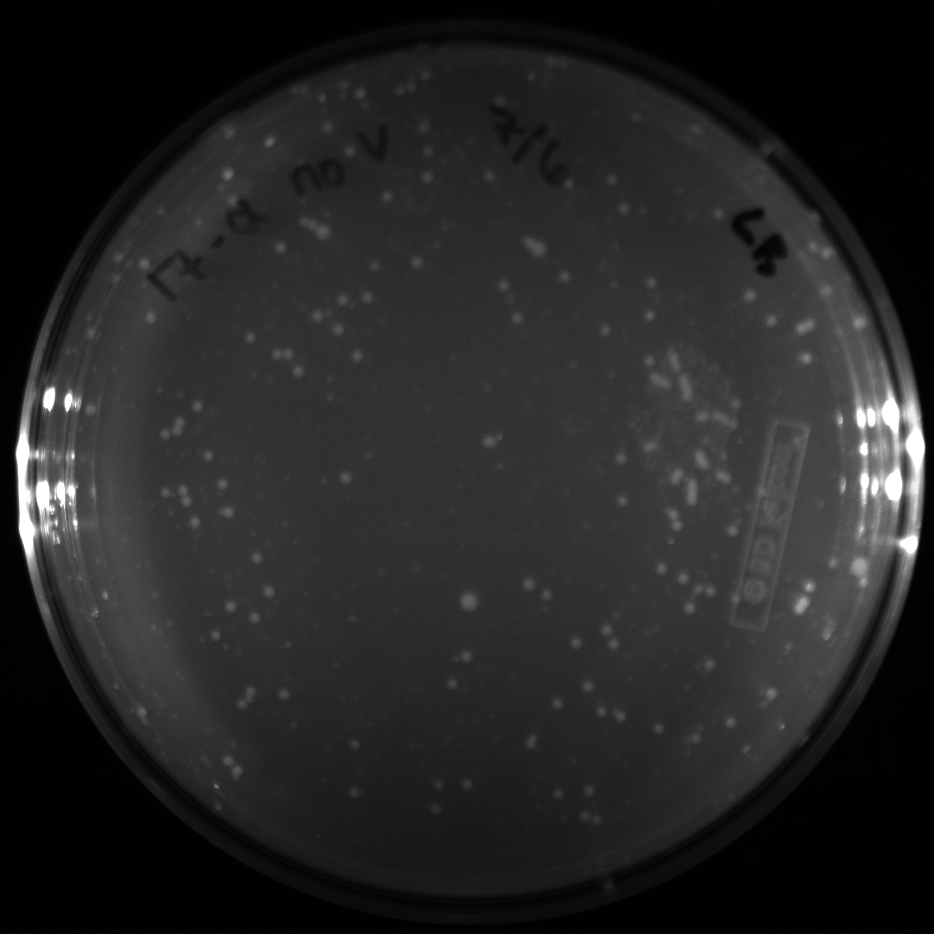
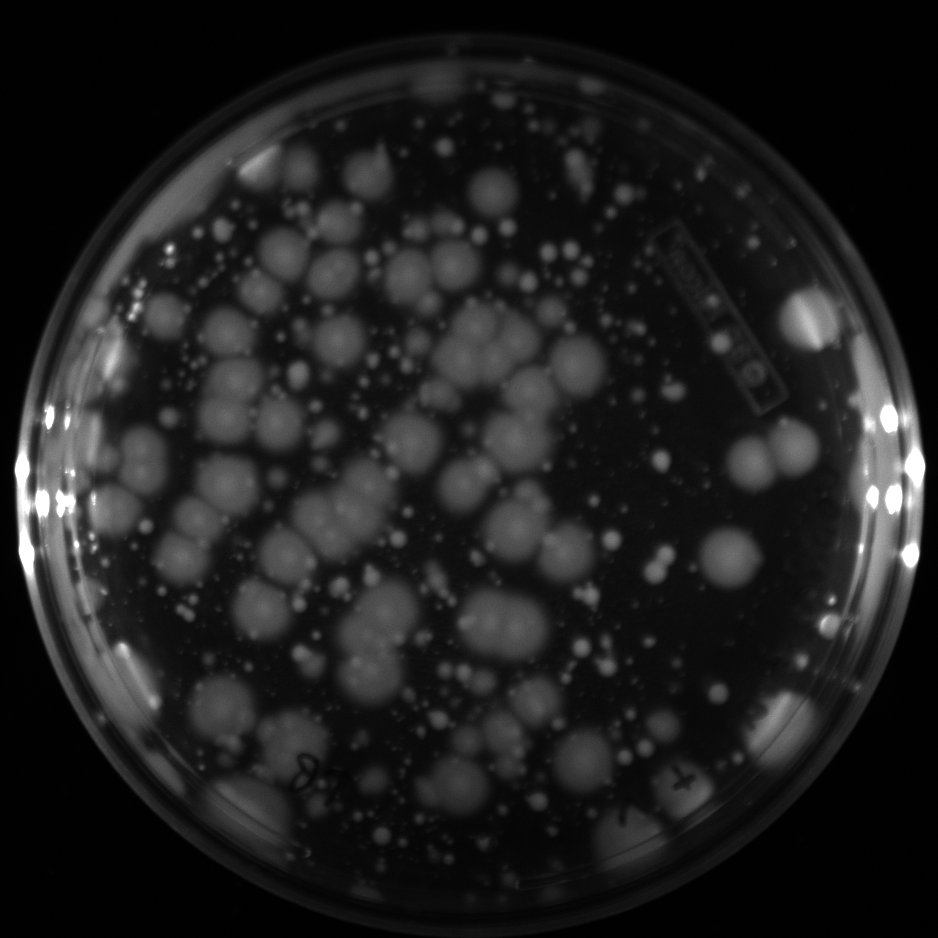
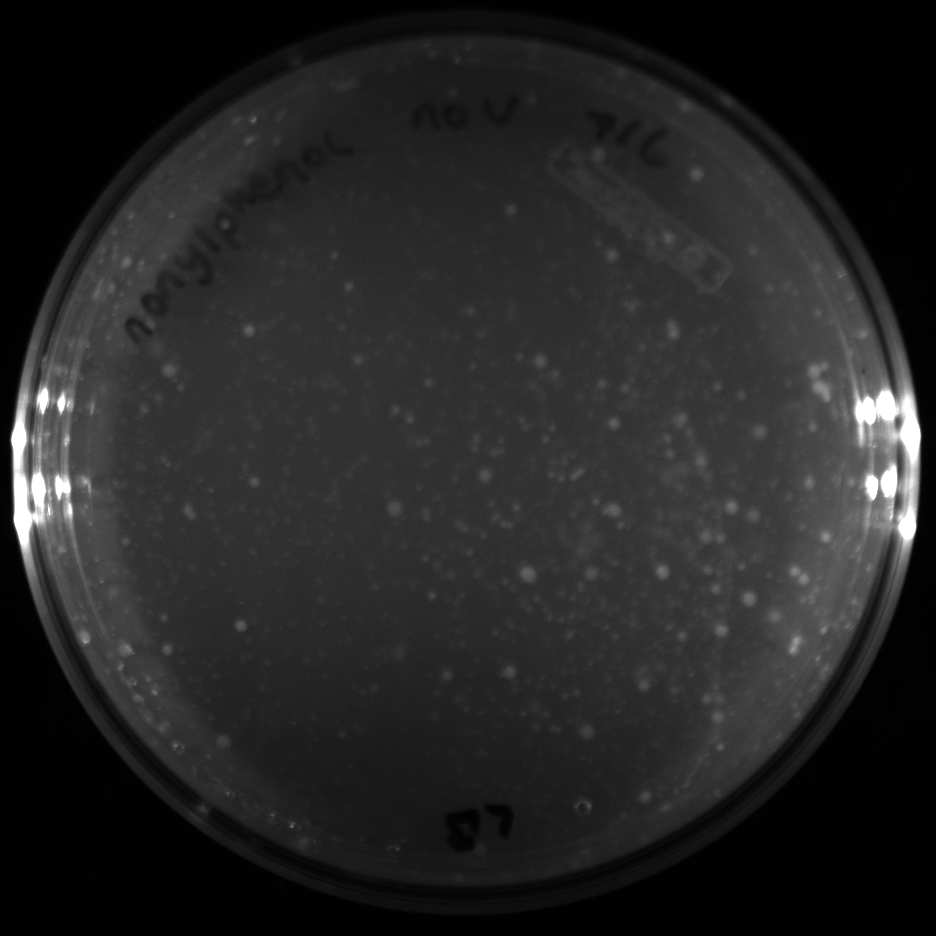
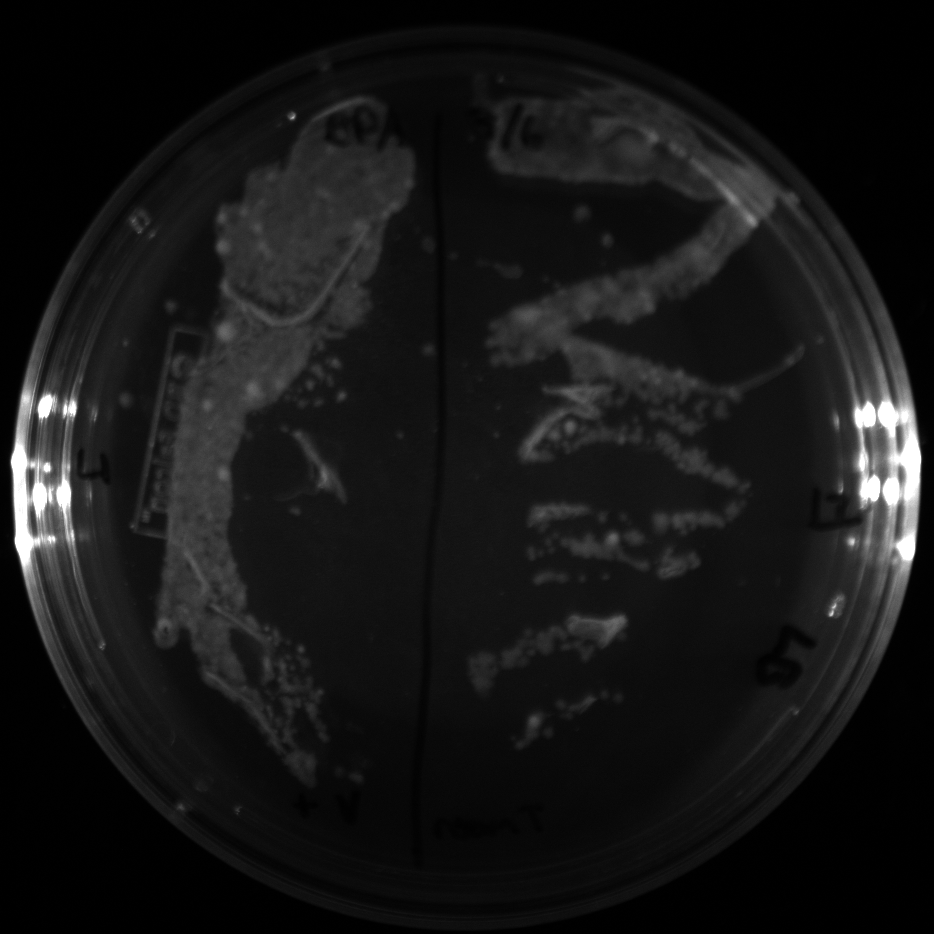
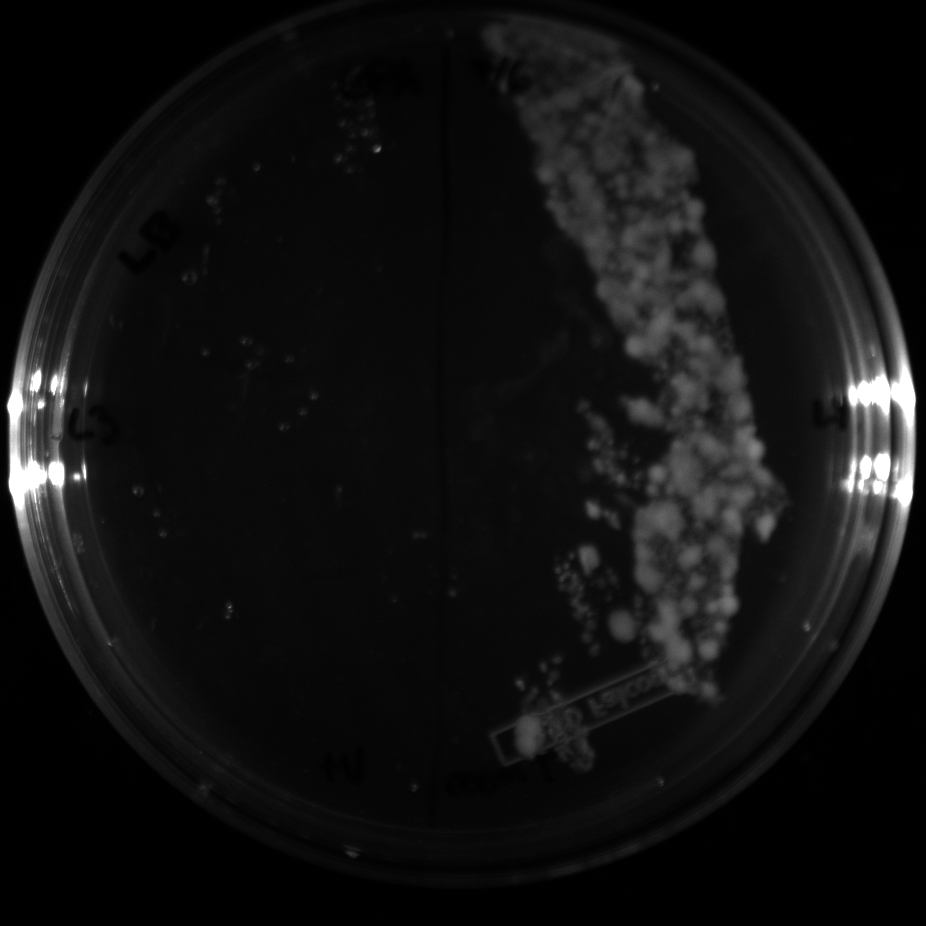
| + | All of the plates of the enrichment cultures showed growth. | ||
| + | <gallery caption="LB Plates of Enrichment Cultures"> | ||
| + | File:Ddtv.png|DDT with vitamins | ||
| + | File:Ddtnov.png|DDT without vitamins | ||
| + | File:Ethynylestradiolv.png|17a-ethynylestradiol with vitamins | ||
| + | File:Ethynylestradiolnov.png|17a-ethynylestradiol without vitamins | ||
| + | File:Nonylphenolv.png|Nonylphenol with vitamins | ||
| + | File:Nonlyphenolnov.png|Nonylphenol without vitamins | ||
| + | File:Bpavroom12.png|BPA with vitamins, room temperature, locations 1 and 2 | ||
| + | File:Bpavroom34.png|BPA with vitamins, room temperature, locations 3 and 4 | ||
| + | File:Bpanovroom12.png|BPA without vitamins, room temperature, locations 1 and 2 | ||
| + | File:Bpanovroom34.png|BPA without vitamins, room temperature, locations 3 and 4 | ||
| + | File:Bpav3012.png|BPA with vitamins, 30˚C, locations 1 and 2 | ||
| + | File:Bpav3034.png|BPA with vitamins, 30˚C, locations 3 and 4 | ||
| + | File:Bpanov3012.png|BPA without vitamins, 30˚C, locations 1 and 2 | ||
| + | File:Bpanov3034.png|BPA without vitamins, 30˚C, locations 3 and 4 | ||
| + | </gallery> </p> | ||
| + | |||
| + | ==July 8== | ||
| + | <p>Send off pNT001 and pNT002 for sequencing again <br/> | ||
| + | Talk to Kenny about PFGE<br/> | ||
| + | Work on design of test constructs for estrogen receptor</p> | ||
| + | |||
| + | ==July 10== | ||
| + | <p>Start overnight culture of pSB4A5 for use in construction of pNT003</p> | ||
}} | }} | ||
Latest revision as of 00:11, 12 July 2011
|
Project |
July 3Checked Gibson transformations ResultsThere was no growth on the transformed Gibson plates. Since we did a small reaction, we used all of the reaction in transforming competent cells. The Gibson assembly likely failed, as one of the negative control plates had a single colony. However, we have had trouble with our competent cells in the past and will try plating what we have left.
July 4ResultsAfter plating the remaining 450 ul of our transformed cells:
July 5Start overnight cultures of Gibson transformations ResultsThe gel of the river DNA was inconclusive, as no bands were visible (even the weight ladder). The gel will be rerun. July 6Miniprep Gibson transformations and send off for sequencing ResultsAgain, no bands were visible on the gel. We will run the samples on a smaller gel to check the size of the DNA and confirm that there is in fact DNA in the sample. July 7Miniprep new biobricks and send off for sequencing ResultsThe smaller gel shows a smear at the top of the gel for one of the samples, indicating that this sample contains DNA that is probably of the correct size for work with the fosmid kit. All of the plates of the enrichment cultures showed growth. July 8Send off pNT001 and pNT002 for sequencing again July 10Start overnight culture of pSB4A5 for use in construction of pNT003
|
 "
"