Team:Arizona State/Project/Software
From 2011.igem.org
|
|
We have developed a tool to assist in the development of synthetic CRISPR systems.
CRISPRstudio is provided "as is" with no express or implied warranty for accuracy or accessibility. Downloads
Installation Using the provided binaries is the fastest way to get started with CRISPRstudio. Visit our GoogleCode page and select the downloads tab. Download the appropriate build based on your OS. Builds are provided using cx_Freeze. All builds are provided for x86 platforms. For real x64 support, you will need to run the source directly. x64 support has not shown significant speed increases for CRISPRstudio. Extract the compressed file to some location. You also need to download BLAST+ 2.2.25 or compatible. BLAST+ must be installed and included in the system path. On most systems, the BLAST+ installer will take care of this. After this, you may run MainFrame to start the program. Source
Pick os/arch as needed. Note that each of the python packages should list the version of Python they support - select the right one. NumPy must be installed before BioPython. The x64 release of wxPython doesn't seem to like Win7 so it may be better to use the x86 release. BLAST+ must be included in the system path. If BLAST+ is not available in the the system, the spacer creator will not function. The information browser will still function normally. The code itself is not fully documented. However, the code was intentionally written to be readable without comments. Project files are available for PyCharm 1.6. You may then run MainFrame.py to start the program. Interface All screen shots were taken on Windows 7, however, CRISPRstudio was designed with portability in mind and will look similar on other operating systems.
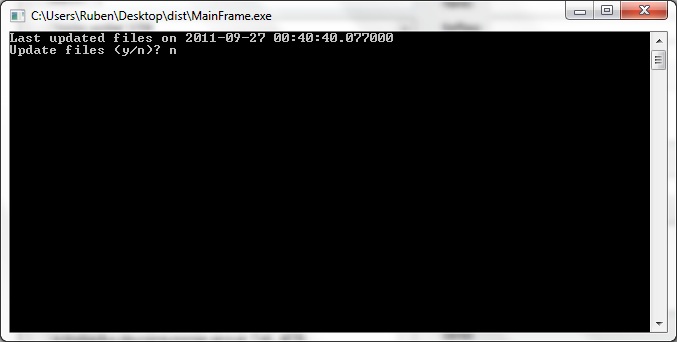 When first launching CRISPRstudio, users are greeted with the above screen. This screen prompts the user to update the local database of CRISPR information. Press y to update or n to continue without updating. An internet connection is required. Note that we directly interface with several online databases and changes in their formats may cause our updater to fail. In this case, the data provided with CRISPRstudio may be used. Release packages of CRISPRstudio include data that was current at time of release.
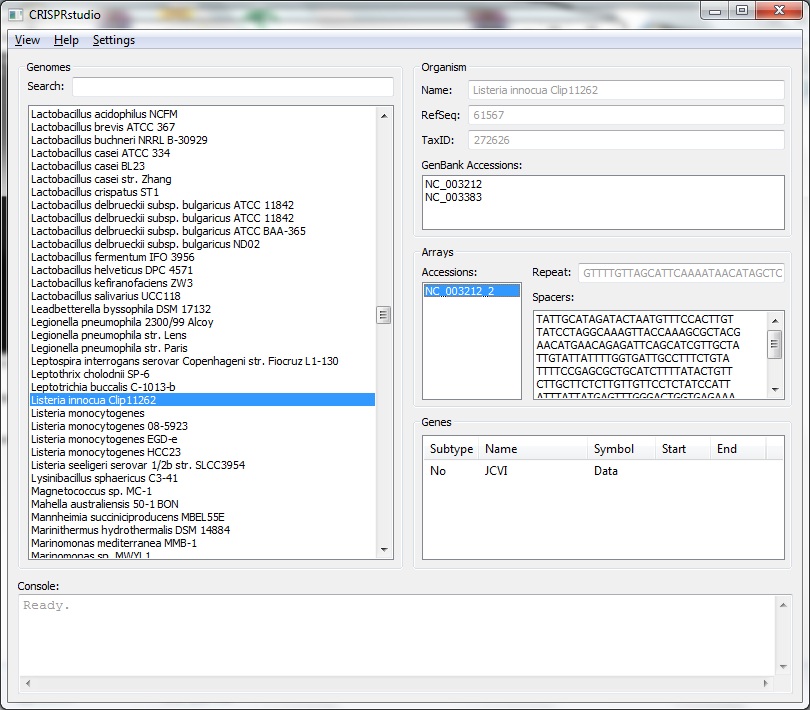 The genomes list is located on the left side of the screen. All genome listed are know to have some form of CRISPR. You may search for a specific genome by entering part of its name in the search bar. Logical connectives may also be used, e.g. entering lactobacillus and bl23 will return Lactobacillus casei BL23. On the left side of the screen is information relating to that genome. We provide basic information about the organism including name (as given by NCBI), RefSeq, Taxonomy ID, and associated GenBank Accessions. For all genomes, we provide a list of arrays with their Accession. For each arrray, we provide the consensus repeat and a list of the spacers in that array. For some genomes, those available in JCVI, we also list the CRISPR genes that known to exist in the genome. Accessions, spacers, and genes may be copied using Ctrl+C on Windows.
 In the upper right side of the screen, you will see a menu bar. The view menu allows the user to switch between viewing CRISPR information and generating spacers. The settings menu controls how spacers are generated. A help menu that provides no actual is also available for piece of mind while using the software.
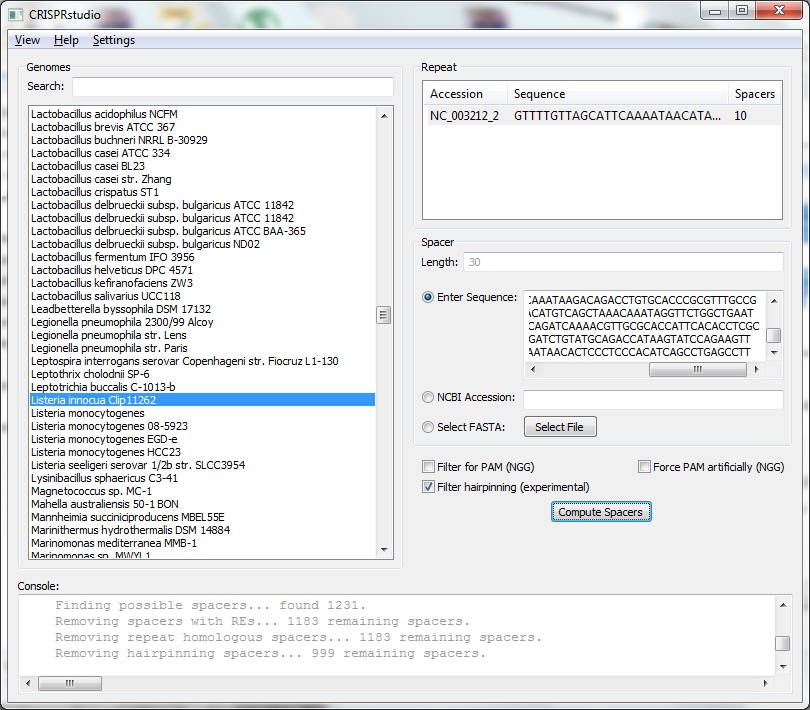 As with the information panel, the genomes list is located on the left side of the screen. The left side of the screen has information for specifying what how to generate spacers. For the selected genome, a list of the arrays are shown based on the repeat that is associated with them. One repeat must be selected. Once a repeat has been selected, the spacer length for that particular array is shown. The user must then enter sequence data for the target of the spacer. A raw DNA sequence may be pasted in, an NCBI Accession entered (with version number), or a FASTA file may be selected. Three additional options are provided for advanced users. The user may choose to Filter for PAM, this discards all potential spacers that do not have the appropriate GG suffix. Typically almost all spacers will be discard if this option is selected. The user may also chose to Force PAM artificially, this will manually inserted the required suffic into all spacers. We also support a basic form of checking for hairpinning of spacers. Depending on the use of the spacer data, it may be more effective to use another tool to check for the existence of hairpinning. After selecting the required settings, the user must click Compute Spacers. This process may take some time depending on the length of the input sequence. An internet connection may be required. During this time, the genome and other sequence information will be automatically downloaded if it does not already exist. We use the following basic process for determining spacers:
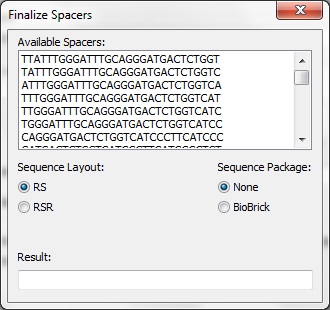 Once CRISPRstudio is done computing the possible spacers, the spacers must be finalized. A list is displayed containing the possible spacers. The user may select how to layout the sequence: repeat-spacer or repeat-spacer-repeat. The user may also chosen to package the spacer using the standard Biobrick prefix and suffix. The final result is display in a textbox at the bottom.
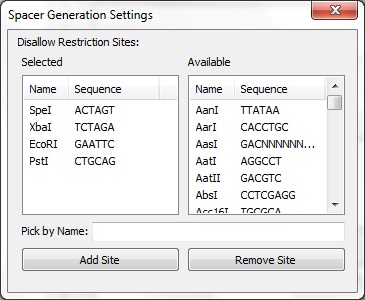 The Spacer Generation Settings dialog controls which REs are considering when creating spacers. The spacer creator requires that a set of REs does not exist in potenial spacers. The set of REs is defined here. On the left is a list of currently selected REs, these may be removed by selecting an item and clicking Remove Site. On the right is a list of all REs known in BioPython. Any of these may be added by selecting on it and clicking Add Site. Alternatively, the name of a RE (e.g. EcoRI) may be entered in the text box and adding by clicking the button. The selected REs are stored locally and will be loaded next time CRISPRstudio is run. Data Sources |
 "
"
