Team:Harvard/Bioinformatics
From 2011.igem.org
Terminology
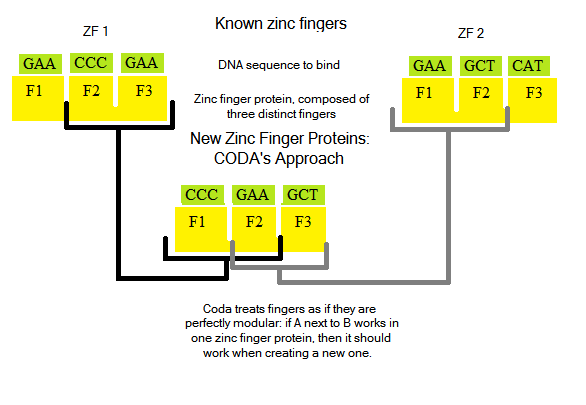
Fingers: made of approximately 30 amino acids, 7 of which form a helix that binds to a DNA triplet Zinc finger proteins (ZFPs): arrays of three fingers that bind to 9 bases (3 triplets) of DNA.
[Diagram]
Past Zinc Finger Designers
Designing new zinc finger proteins (ZFPs, which are arrays of three fingers) is not an easy task: how they bind and interact with DNA bases is not fully understood, and is an active area of research [Persikov]. Notable past attempts to create novel ZFPs [CODA, OPEN] tried a two distinct methods: CODA took a modular approachOPEN took two known fingers from an array, and randomized protein sequences to try to generate a third finger to bind a new triplet:
Both techniques were successful in finding ZFPs to bind to novel DNA sequences [how successful?]
Our Approach
Improving on the concept of OPEN, we decided to design ZFPs where the first two DNA triplets can be bound, but the third cannot. For example, if the sequence GTG GGA CCA can be bound but GTG GGA TGG cannot, we would use the first two fingers and generate the third. OPEN simply randomized amino acid sequences to try to create a third finger
 "
"