Team:Paris Bettencourt/Experiments/T7 diffusion
From 2011.igem.org

Experiments - T7 RNA polymerase diffusion
The T7 RNA polymerase diffusion design was the first one we thought of. It uses the RNA polymerase of the T7 phage, giving us perfect orthogonality to B.subtilis genome. We use an auto-amplification device base on this polymerase that we call T7 autoloop. This autoloop is also used in the tRNA amber diffusion design. We successfully BioBricked and characterized all our constructs for this design and made them available for the synthetic biology community. Our results are detailed below.
Abstract
Results for the YFP concentrator:
- We successfully BioBricked both the T7 RNA polymerase emitter (BBa_K606039) and the T7 autoloop (BBa_K606036) constructs and sent them to the registry
Design overview

Schematic summary of the T7 diffusion device
All of the parts of the above design have been BioBricked, characterized both in E.coli and B.subtilis, and sent to the registry.
More details on the design are available here.
Parts and BioBricks construction
You can find the cloning plan for the T7 RNA polymerase design below:
INSERT CLONING PLANSetback with the pHyperspank promoter
At the beginning of the project, we intended to use a pHyperSpank promoter (Ph-s, BBa_K143055) in front of the emitter construct. The Ph-s promoter is repressed by LacI and would have allowed us to use an IPTG-inducible promoter. However, our first experiments with the promoter were inconclusive. We sequenced our sample and realized that we had received a BioBrick of the size of Ph-s but not the one we expected. It was anoter gene entirely. Rather than losing weeks of experiments waiting for another HyperSpank promoter, we chose to use a constitutive Pveg promoter (BBa_K143053).
Characterization of the T7 promoter
In order to characterize the pT7 promoter, we used the construct pT7-RBS-GFP-T7ter. This construct was transformed into BL21 strains (E.coli) expressing the T7 polymerase under IPTG induction.Fluorescence kinetics
The measurements have been carried out on a spectrophotometer at 37°C under transient shaking. The experiment lasted 4h, we tested several colonies and several IPTG concentrations. The OD at 600nm and the fluorescence of the GFP (exc: 470nm / meas:515 nm) was measured every 5 min and the ratio of the two was calculated.
All values were normalized by substracting the fluorescence/OD value of the well with 0 mM IPTG at time 0. The values given are in arbitrary units.
After 2 hrs of induction, we see a clear increase of the fluorescence proportional to the IPTG concentration (that is to say with the quantity of T7 polymerase induced in the cell). After 4 hrs, the expression of GFP under the pT7 is still not saturated
Here we plot the ratio of induction of the T7 polymerase dependant construct for the different concentrations of IPTG at a given time (4 hrs) taking the well with 0 IPTG at time 0 as the reference.
Characterization of the T7 RFP emitter <--TO BE CORRECTED HEAVILY
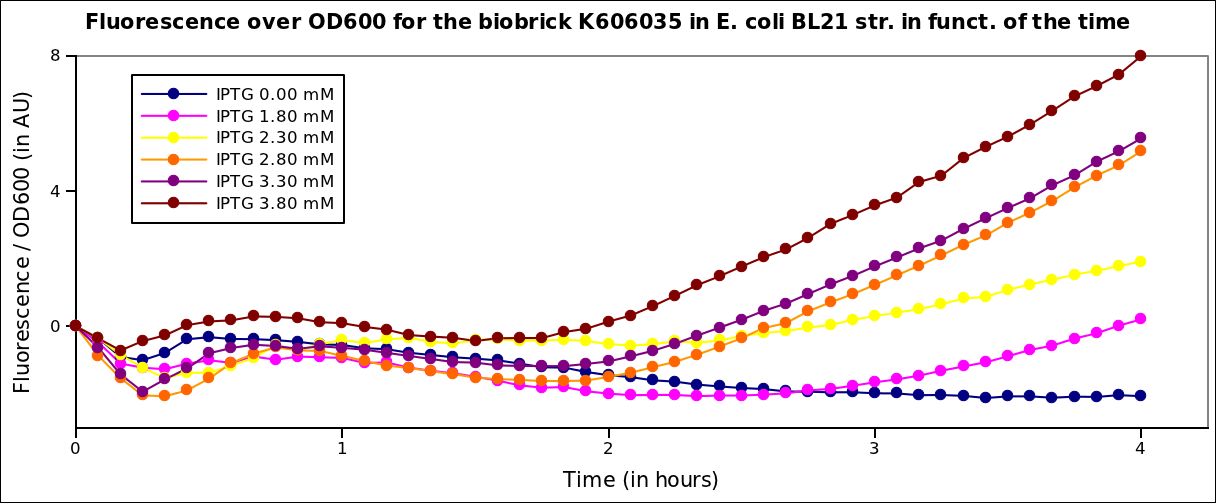
We found out there is a small leakage with the T7 emitter RFP. The cells in which RFP production is activated stop dividing and glow very strongly.
The first pictures show that the RFP construct is working efficiently since some cells are glowing with RFP fluorescence. This also shows the system is not as leaky as we expected. Indeed, the promoter regulating RFP expression is 'pVeg' which is a constitutive promoter. Finally, the RFP system is working very well because when leak occurs, cells glow very strongly.
Characterization of the T7 signal amplification leakage
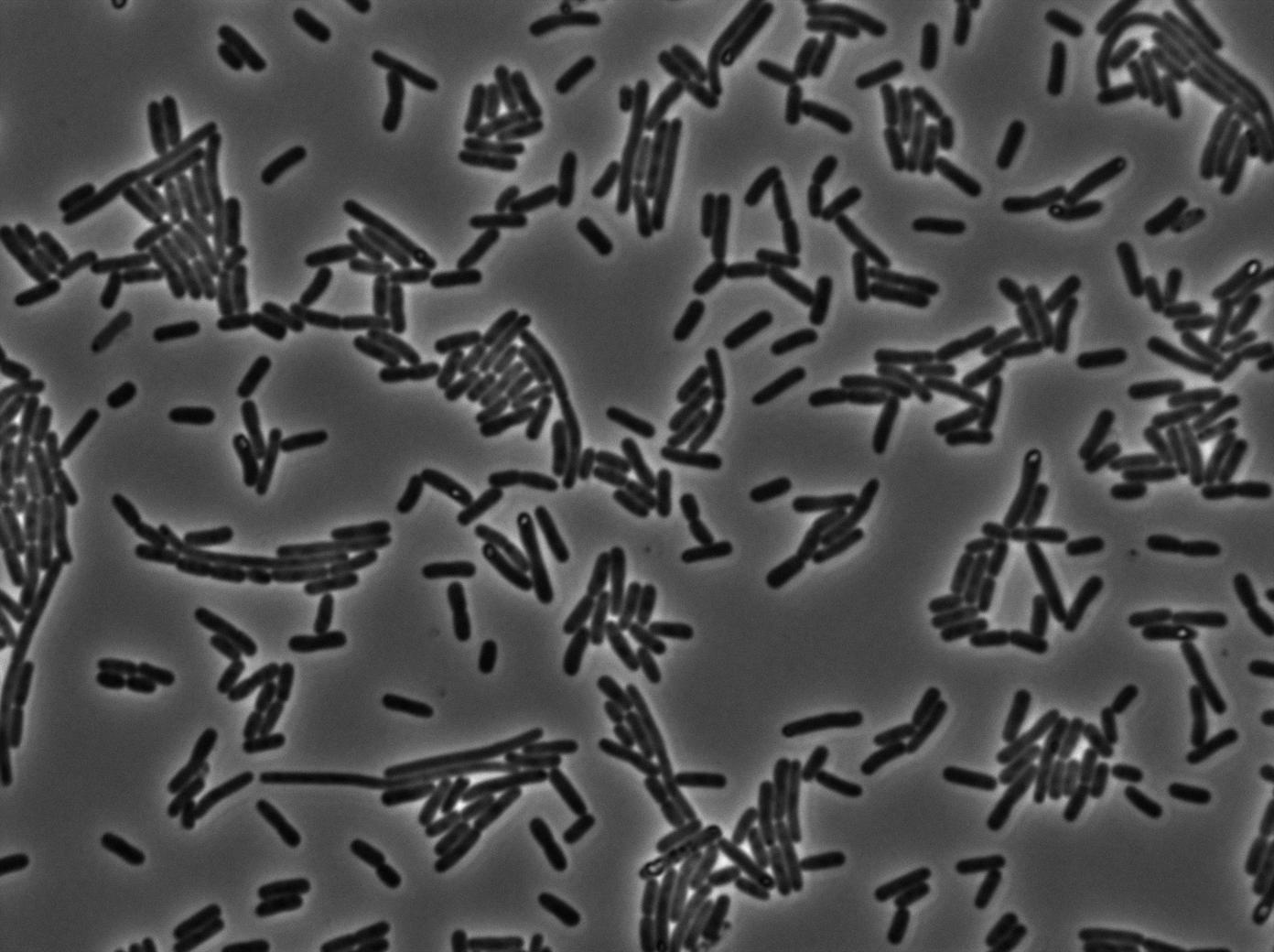
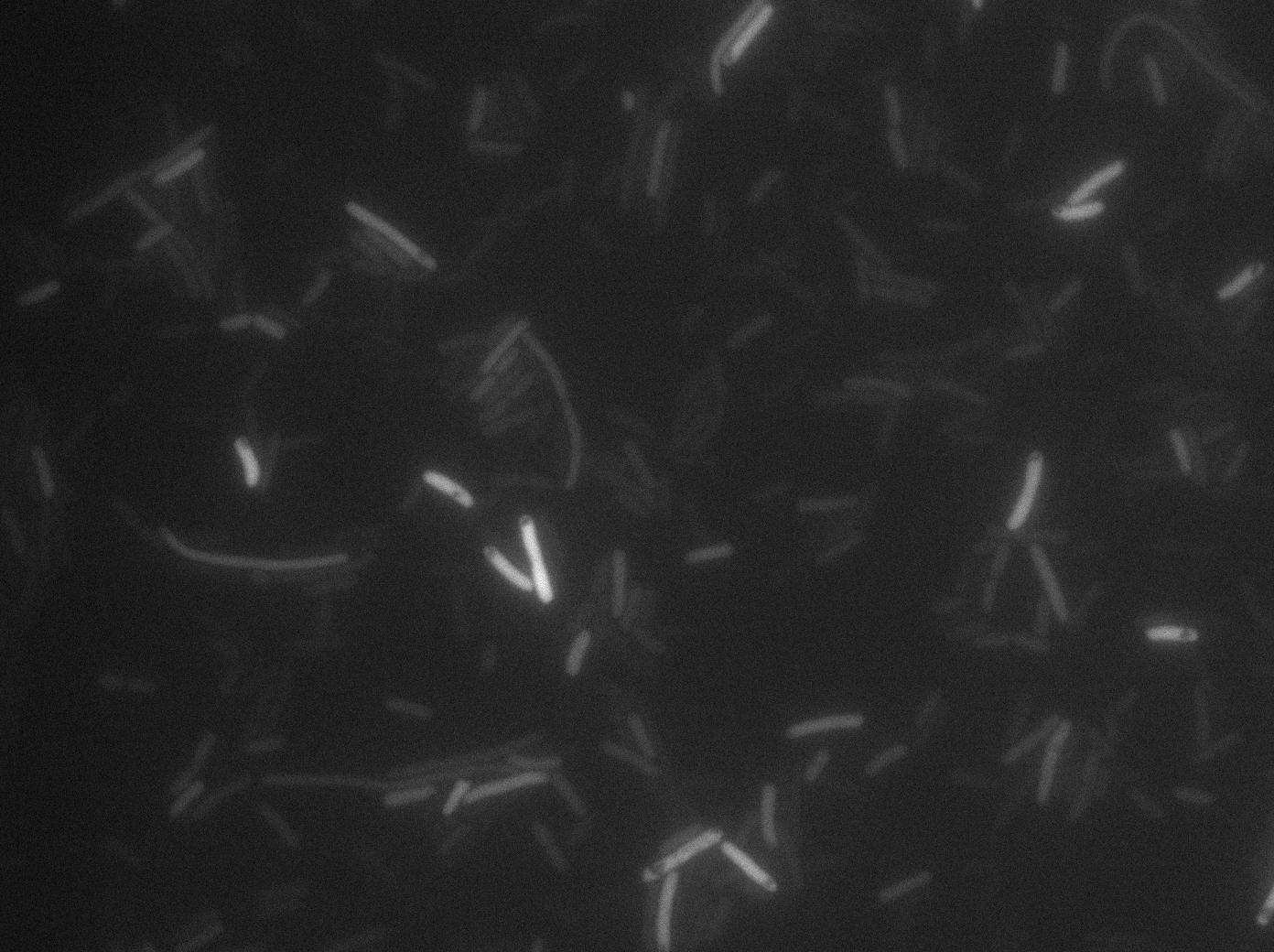
We characterized T7 autoloop (receiver part of the construct, BBa_K606036) in E.coli, when hosted in the plasmid pSB1C3. The idea was to see if the system was leaky inside a synthetic biology plasmid, that is to say, a plasmid holding 4 terminators before any construct.
These pictures show that the T7 GFP autoloop system is efficient since some cells are glowing with GFP fluorescence. Thus, we can conclude that the T7 polymerase is activated. Finally, we notice there is no difference between the GFP autoloop with and without terminator before the T7 promoter. It could be due to that terminator which is a B.subtilis terminator. Moreover, we know that this E.coli strain has 4 terminators.
 "
"