Team:NTNU Trondheim/Data
From 2011.igem.org
m |
(→Data) |
||
| Line 3: | Line 3: | ||
= Data = | = Data = | ||
| + | |||
| + | == rrnB P1 promoter == | ||
| + | |||
| + | The rrnB P1 promoter is negatively regulated by ppGpp [Kilde]. We found the | ||
| + | promoter in the registry distribution (BBa_K112118) submitted by Berkly in 2008. | ||
| + | |||
| + | This part is in the BBb standard form [http://openwetware.org/wiki/Template:AndersonLab:BBb_Standard] | ||
| + | and is not compatible with the parts in the BBa standard. In order to get the part in BBa standard we | ||
| + | PCR amplified the promoter using the BBa_K112118 as template and primers containing the BBa prefix and suffix: | ||
| + | |||
| + | {| border="1" | ||
| + | |- | ||
| + | !Primer | ||
| + | !Type | ||
| + | !Sequence | ||
| + | |- | ||
| + | |rrnB P1 F | ||
| + | |Forward | ||
| + | |GTTTCTTCGAATTCGCGGCCGCTTCTAGAGACGTATCCTACGCCCGTGGT | ||
| + | |- | ||
| + | |rrnB P1 R | ||
| + | |Reverse | ||
| + | |GTTTCTTCCTGCAGCGGCCGCTACTAGTACGCCTTCCCGCTACAGAGTCA | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | The PCR-product was run on a 1,5% agarose gel in order to verify that we had achieved the correct product and to separate it from any other unwanted products. | ||
| + | |||
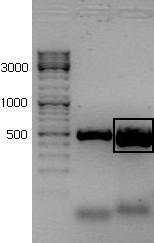
| + | [[File: Primer_rrnBL_060711_resultat.JPG|thumb|PCR products from rrnB P1 BioBrick separated on 1.5 % agarose. The marked products represent rrnB P1 with regular pre/sufix]] | ||
| + | |||
| + | The new rrnB P1 biobrick was inserted in the psb1C3 plasmid provided by the iGem HQ. | ||
| + | Plasmid from five colonies were digested with the enzymes ....... and .............. giving the lengths ........... and ................ if the promoter was inserted. | ||
| + | |||
| + | [[File: Test_rrnB_C3_060811_wiki.jpg|thumb|PCR products from rrnB P1 BioBrick separated on 1.5 % agarose. The marked products represent rrnB P1 with regular pre/sufix]] | ||
| + | |||
| + | It seems like parallel 1 and 2 have the insert. | ||
| + | |||
| + | |||
| + | |||
{{:Team:NTNU_Trondheim/NTNU_footer}} | {{:Team:NTNU_Trondheim/NTNU_footer}} | ||
Revision as of 12:24, 23 August 2011

Data
rrnB P1 promoter
The rrnB P1 promoter is negatively regulated by ppGpp [Kilde]. We found the promoter in the registry distribution (BBa_K112118) submitted by Berkly in 2008.
This part is in the BBb standard form [http://openwetware.org/wiki/Template:AndersonLab:BBb_Standard] and is not compatible with the parts in the BBa standard. In order to get the part in BBa standard we PCR amplified the promoter using the BBa_K112118 as template and primers containing the BBa prefix and suffix:
| Primer | Type | Sequence |
|---|---|---|
| rrnB P1 F | Forward | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGACGTATCCTACGCCCGTGGT |
| rrnB P1 R | Reverse | GTTTCTTCCTGCAGCGGCCGCTACTAGTACGCCTTCCCGCTACAGAGTCA |
The PCR-product was run on a 1,5% agarose gel in order to verify that we had achieved the correct product and to separate it from any other unwanted products.
The new rrnB P1 biobrick was inserted in the psb1C3 plasmid provided by the iGem HQ. Plasmid from five colonies were digested with the enzymes ....... and .............. giving the lengths ........... and ................ if the promoter was inserted.
It seems like parallel 1 and 2 have the insert.
 "
"