July 31
Started a LB+amp overnight culture of the B0034 glycerol stock from the 2010 Caltech iGEM Team
Results
Mass spectrometry did not run correctly; samples did not ionize; must rerun
August 1
PCR pSB3K3 and pNT003 insert with the new primers.
Gel extract the pNT002 insert from the July 29 PCR
Standard Assembly Digest: K123001, B0014, B0034, R0010, pSB4A5
Minimal media transfers
Transformation and plating of pUC19 into competent cells for cell dry weight experiment
Overnight culture of B0034 glycerol stock
Overnight ligation of pNT002 insert and PSB3K3 backbone
Restriction digest of parts for pNT002 standard assembly and overnight ligation
Results
No colonies visible on the pUC19-transformed cells
The enrichment culture plates show no visible growth yet.
Gel showed that the terminator, RBS, and lac promoter did not digest for standard assembly.
Gel Extractions of July 29 PCR of pNT002 and pNT003 inserts
| Part |
Concentration (ng/ul) |
| pNT002 insert |
65.9 |
| pNt003 insert |
12.5 |
August 2
Gibson assemble pNT002 using the gel extracted insert and linearized pSB3K3
Gibson assemble pNT003 using the gel extracted insert and linearized pSB4A5
Transform Gibson reactions into our XL-10 cells and Emzo's top ten cells
Replating of more pUC19-transformed competent cells and non-transformed competent cells
Standard Assembly Digest- K12300, B1004, R0010, B0034, along with the destination plasmids.
Run gel of overnight pNT002 ligations
Transformed pNT002 insert+ pSB3K3 ligation
Results
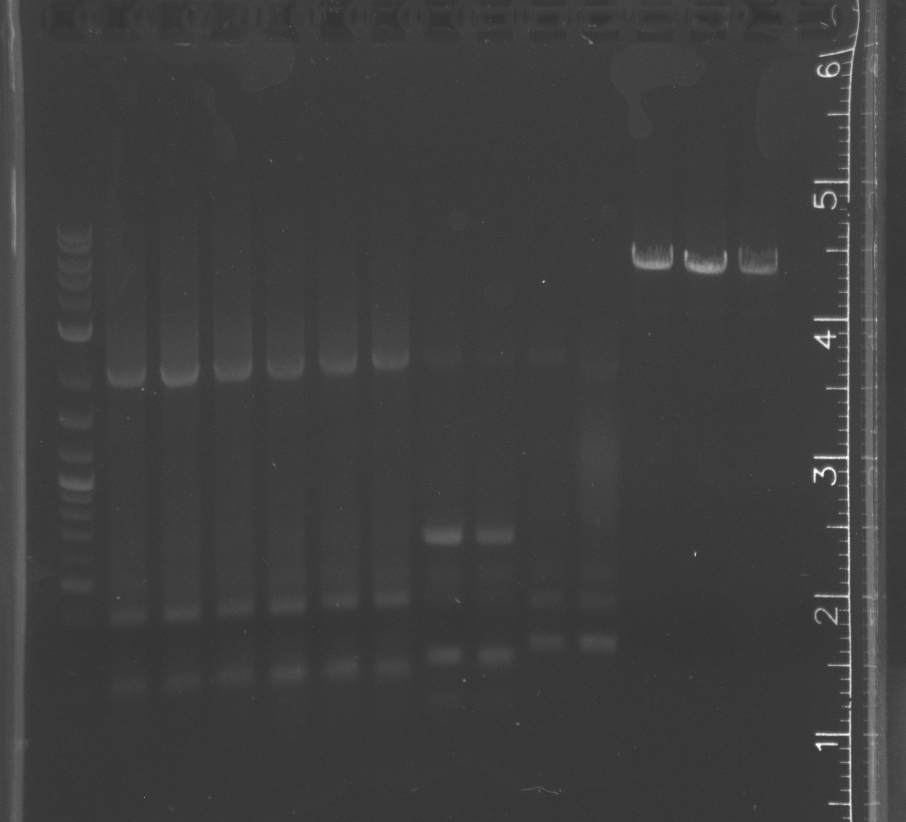
lane 1 NEB 2-log ladder, 2-4 pNT003 insert, 5-7 pNT003 insert with PIPE primers, 8-13 pSB3K3, 14-15 pETDEST53
|
lane 1 NEB 2-log ladder, 2-4 pNT003 insert, 5-7 pNT003 inset with PIPE primers, 8-9 pNT003 insert, 10-11 pNT003 inset with PIPE primers, 12-15 pETDEST53
|
pNT002 ligation showed band on gel
| Part |
Concentration (ng/ul) |
| pSB3k3-1 |
23.8 |
| pSB3K3-2 |
29.9 |
| pETDEST53 |
90.9 |
August 3
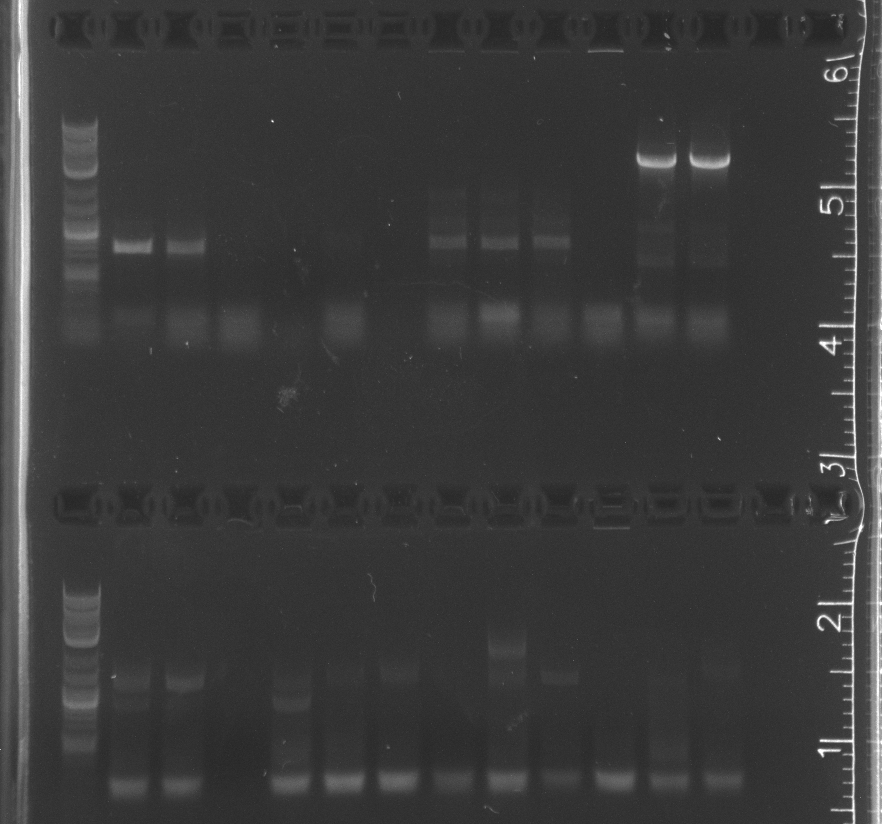
Colony PCR of Gibson transforms
Another assembly digest with gel extraction.
Emzo's protocol for traditional assembly of PCR'd pNT002 insert and pSB3K3 plasmid
Results
| Plate |
number of colonies |
| pNT002 + top ten |
55 |
| pNT002 - top ten |
43 |
| pNT002 +XL10 gold |
180 |
| pNT002 - XL10 gold |
400 |
| pNT003 + top ten |
14 |
| pNT003 - top ten |
1 |
| pNT003 +XL10 gold |
0 |
| pNT003 - XL10 gold |
7 |
Only the enzyme and the vector had correct band lengths on the gel. We extracted them and nanodropped, but concentrations too low.
| Part |
Concentration (ng/ul) |
| K123001 |
2.4 |
| pSB4A5 |
6.2 |
August 4
Ran gel of colony pcr from yesterday
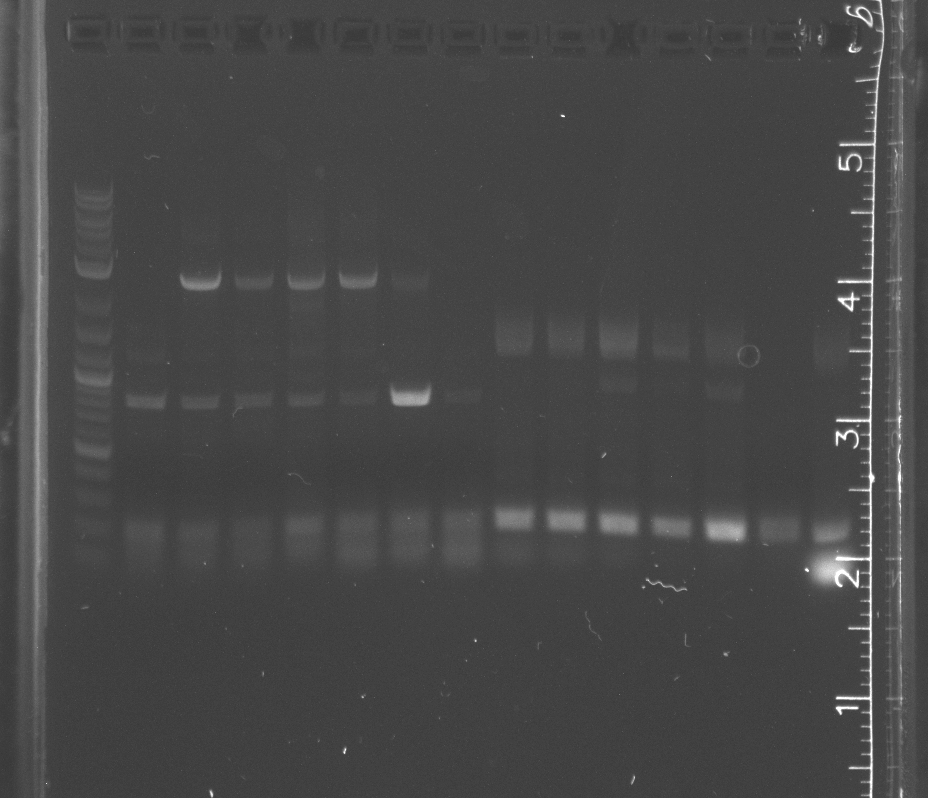
Ran two separate gels for standard assembly according to the number of base pairs each had.
The first 1.3% agarose gel had a 100 bp ladder with R0010 (200 bp), B0014 (95 bp), and B0034 (12 bp).
The second 1% agarose gel had a 2-log ladder with K123001 (1284 bp) and pSB4A5 (3395 bp).
Grow two overnight cultures of 175mL LB-chlor in preparation of dry cell experiment
Results
Some of the colony PCR have insert amplifications of the correct length. Will send some off for sequencing tomorrow.
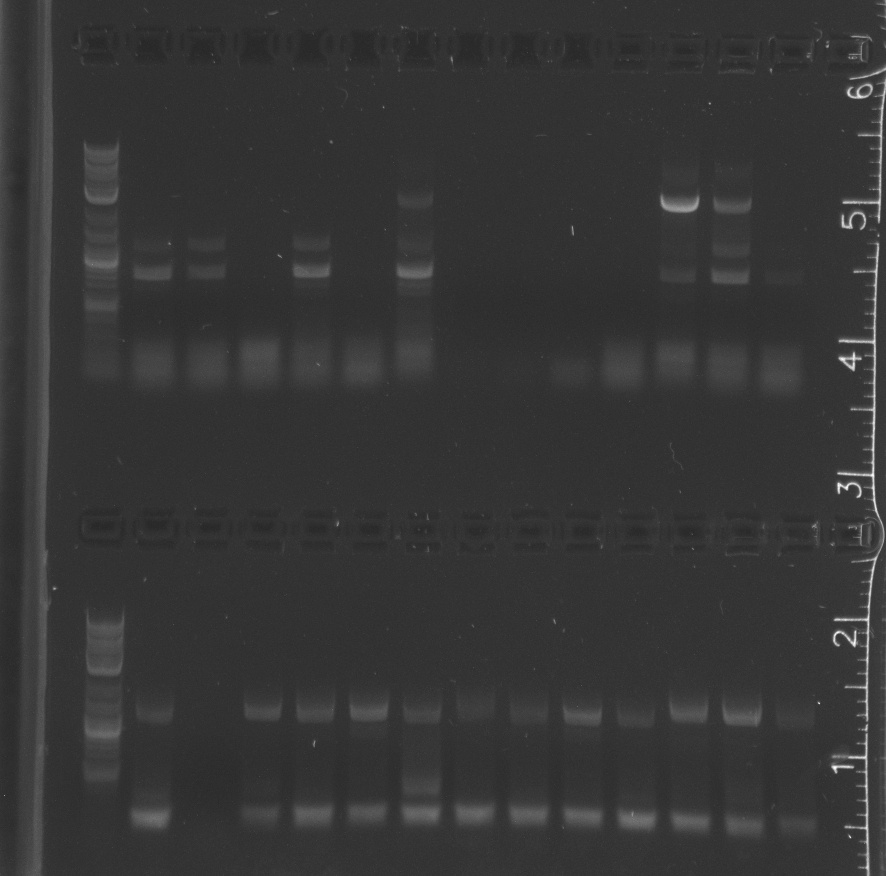
upper comb lane 1 NEB 2-log ladder, 2-14 pNT002 colonies 1-13 vector amplification; lower comb 1 NEB 2-log ladder, 2-14 pNT002 colonies 1-13 insert amplification
|
lane 1 NEB 2-log ladder, 2-8 pNT002 colonies 14-20 vector amplification, 9-15 pNT002 colonies 14-20 insert amplifications
|
upper comb lane 1 NEB 2-log ladder, 2-13 pNT003 colonies 1-12 vector amplification; lower comb 1 NEB 2-log ladder, 2-13 pNT003 colonies 1-12 insert amplification
|
No growth from traditional assembly on self ligation control or experimental plate
Blue light showed bands for all the parts, but again the smaller parts had the wrong band lengths.
August 5
Miniprepped colonies from Gibson colony pcr (pNT003-2,8,11,12 and pNT002-6,11,12,14)
Sent pNT002-14 and pNT003-2,8,11,12 off for sequencing as the others were too dilute
PCR 16s sequences
Rerun p450 degradation assay for GCMS analysis
Results
| Miniprep name |
Concentration (ng/ul) |
| pNT003-2 |
45.3 |
| pNT003-8 |
32.3 |
| pNT003-11 |
33.1 |
| pNT003-12 |
33.1 |
| pNT002-6 |
9.2 |
| pNT002-11 |
12.4 |
| pNT002-12 |
12.1 |
| pNT002-14 |
78.1 |
The gel for 16s had no bands in any of the control or sample lanes.
 "
"