Team:Paris Bettencourt/tRNA diffusion
From 2011.igem.org
(Difference between revisions)
m (html) |
m |
||
| Line 5: | Line 5: | ||
<h1>The tRNA amber diffusion</h1> | <h1>The tRNA amber diffusion</h1> | ||
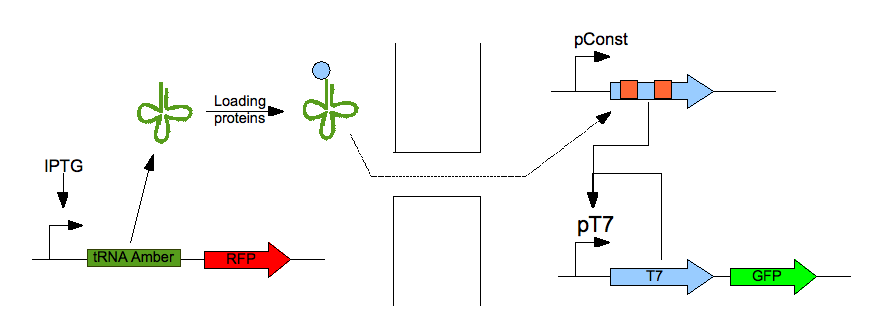
| - | <p>What we call a <em>tRNA amber</em> is a transfer RNA which anticodon | + | <p>What we call a <em>tRNA amber</em> is a transfer RNA which anticodon has been modified for it to be complementary with a stop codon and especially with an amber stop codon (UAG). Our idea was to use the nanotubes to allow a receiver cell to translate an mRNA containing an amber mutation. The emitter cell will produce this mutated transfer RNA (we mutated a YtRNA for it to recognise the stop codon) and the receiver cell will then be able to translate a protein (we chose the T7 polymerase) which gene contain an amber mutation. This protein will part of a reporter system.</p> |
</html> | </html> | ||
| Line 12: | Line 12: | ||
[[File:tRNA_Amber_principle.jpg|thumb|center|upright=3.0|The tRNA Amber supressor design principle]] | [[File:tRNA_Amber_principle.jpg|thumb|center|upright=3.0|The tRNA Amber supressor design principle]] | ||
| - | We | + | <html> |
| + | <h1>The tRNA amber system preparation</h1> | ||
| + | |||
| + | The first step of this preparation was to choose the tranfer RNA and the amino acids to modify in the T7 polymerase gene. We found the article of Grundy and Henkin in 1994 that led us to use the YtRNA. We synthetysed it by an elongation of primers. Now that we know which codon we have to modify within the T7 polymerase sequence. We looked for those codon in the T7 polymerase sequence and realised a quick change mutagenesis, on it to modify two codons. Indeed we decided that only one modified codon would be too leaky, therefore we changed two of them and in the beginning of the sequence for the ribosome to still be tightly attached to the mRNA. | ||
| + | |||
| + | |||
<html> | <html> | ||
Revision as of 13:44, 15 September 2011

The tRNA amber diffusion
What we call a tRNA amber is a transfer RNA which anticodon has been modified for it to be complementary with a stop codon and especially with an amber stop codon (UAG). Our idea was to use the nanotubes to allow a receiver cell to translate an mRNA containing an amber mutation. The emitter cell will produce this mutated transfer RNA (we mutated a YtRNA for it to recognise the stop codon) and the receiver cell will then be able to translate a protein (we chose the T7 polymerase) which gene contain an amber mutation. This protein will part of a reporter system.
We summed up this principle in the scheme below:
 "
"