Yeast Toolkit Results
Yeast Vector Platform Results
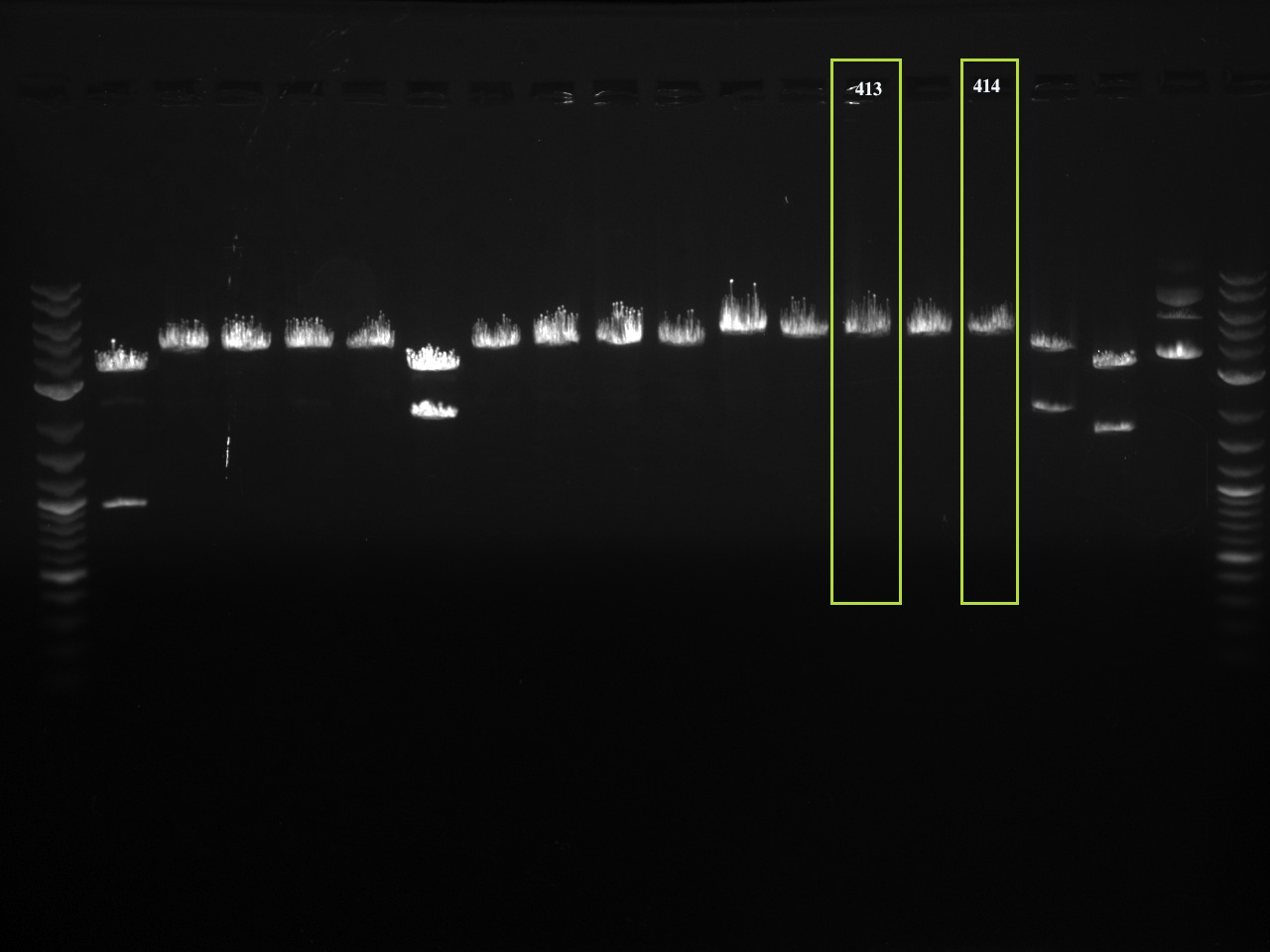
The first set of gels are of restriction enzyme digests corresponding to a restriction enzyme site that was removed using QuickChange PCR. Without the QCPCR, there would be two restriction enzyme sites for the enzyme tested and therefore two products on the gel. The enzyme sites removed all correspond to a standard assembly enzyme: EcoRI, XbaI, SpeI or PstI. With the removal of one of the sites, the cut linearizes the vector, showing one band (and no supercoiled vector). The vector number is above each lane and successful products are squared in green.
The number corresponding to each lane is the pRS vector number. The number code is as follows:
4XY
- If X = 0, it is an integrating vector.
- If X = 1, it is a CEN/ARS vector.
- If X = 2. it is a 2micron vector.
- If Y = 3, it is a HIS3 selectable marker in yeast.
- If Y = 4, it is a TRP1 selectable marker in yeast.
- If Y = 5, it is a LEU2 selectable marker in yeast.
- If Y = 6, it is a URA3 selectable marker in yeast.
Note: all vectors contain Ampicillin resistance for selection in E. coli.
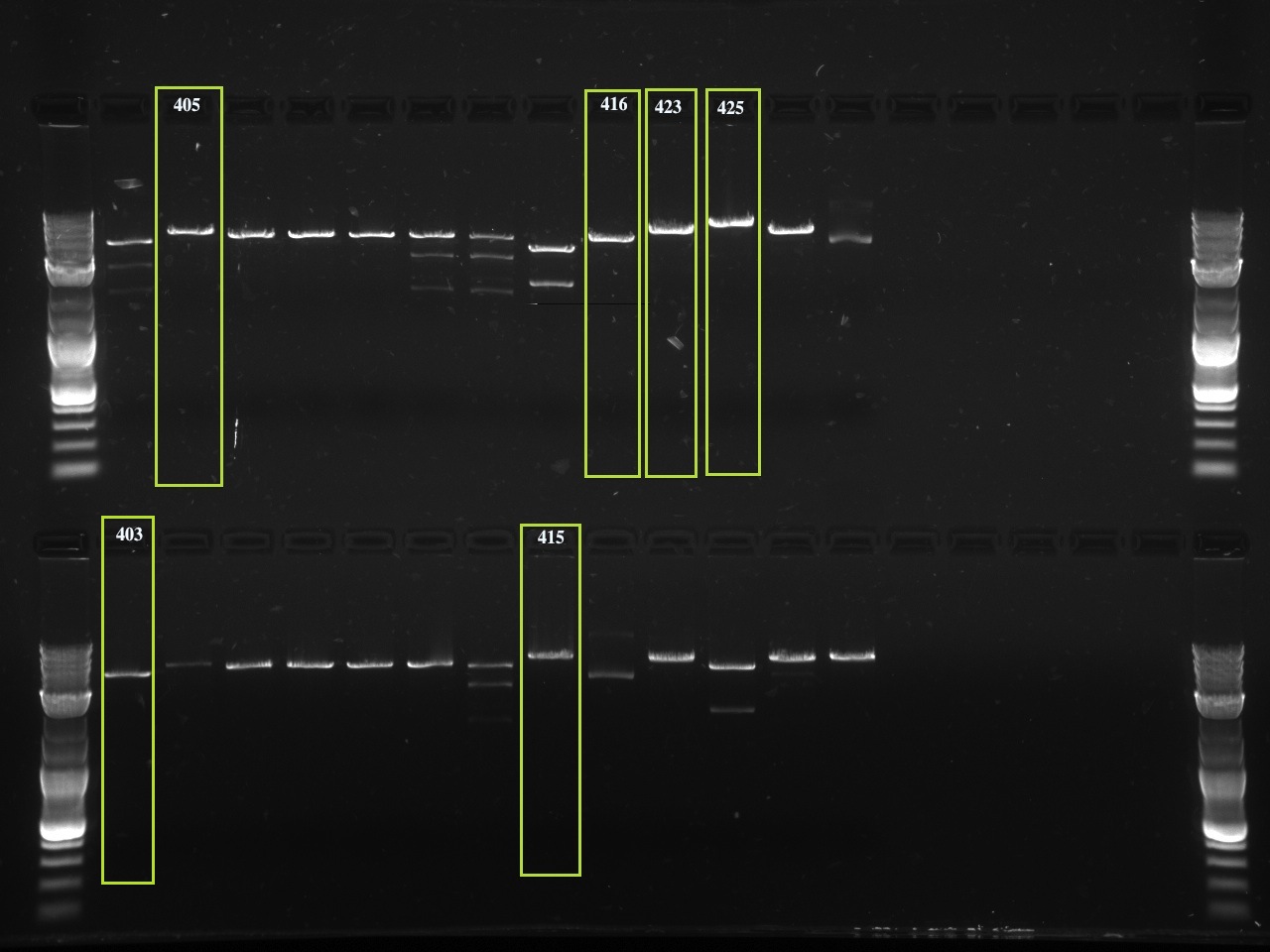
The second set of gels are to ensure the correct BioBrick multiple cloning site was substituted for the original multiple cloning site (the new MCS being in an EcoRI-XbaI-SpeI-PstI digest order) again using QuickChange PCR. The original multiple cloning site contained a XhoI restriction site, while the new BioBrick multiple cloning site does not. Therefore, we digested the new vector with XhoI and compared it to an uncut vector. There are 3-4 samples of each vector tested, for a total of 10 vectors tested. The first lane starts with a XhoI digest and the lane directly next to it is an uncut vector. Therefor the gel alternates between cut and and uncut DNA, with 6 or 8 lanes corresponding to one vector. If a vector was uncut by XhoI, then the correct multiple cloning site is in place. Note that undigested, supercoiled DNA (the major gel product of undigested vector) run further than linearized, cut vector.
Promoter/Terminator Library
We biobricked 66% of the promoters and terminators that we set out to build. This will improve the ability of future igem teams that will use yeast to perform synthetic biology. We have successfully transformed a number of promoters into yeast(Bap2p, Tdh3p, Kre9p, Hh01p, Pry1p) in order to control expression of a GFP gene. The promoters are ready for flow-cytometry analysis.
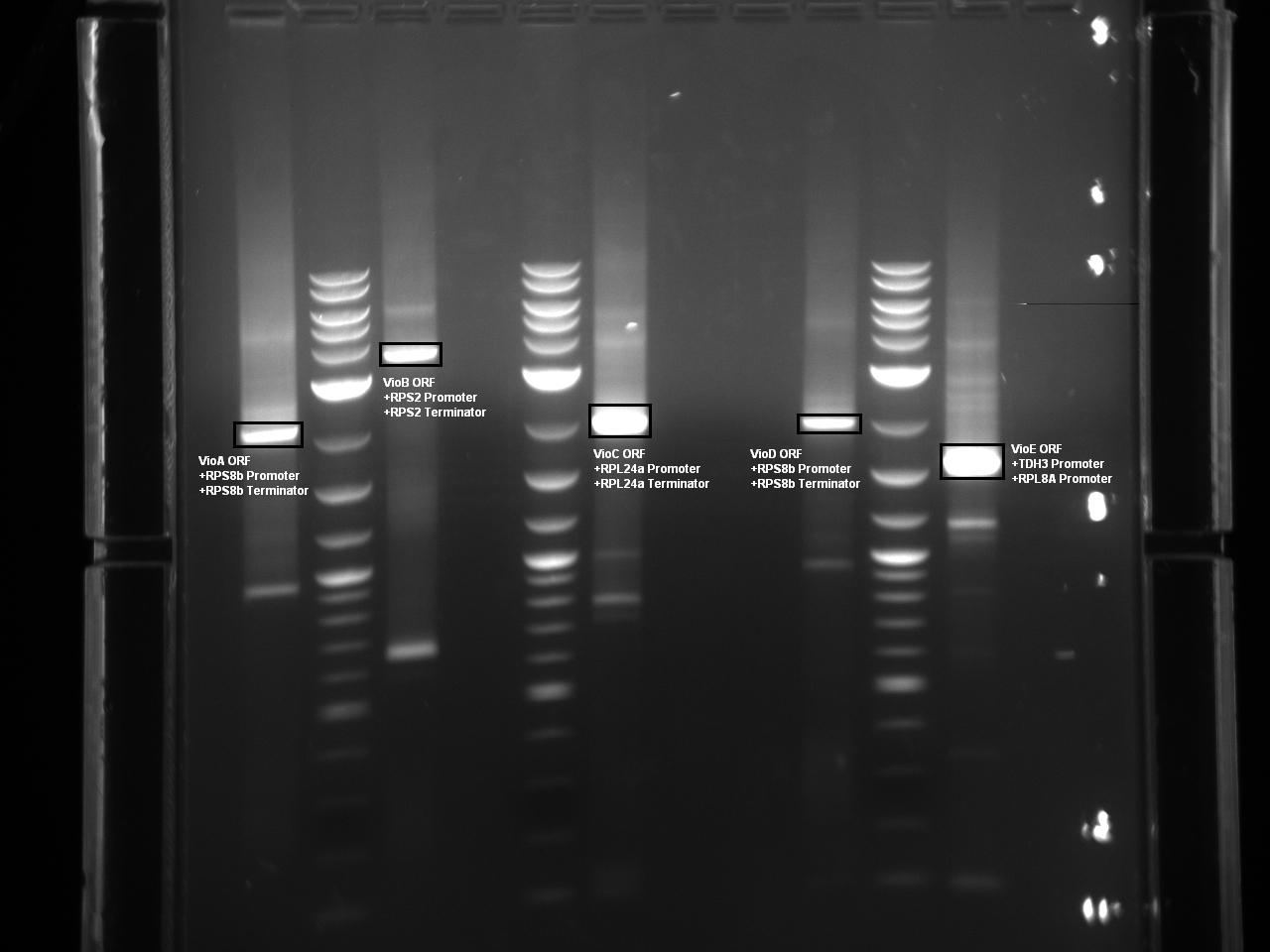
Five Violacein Expression Cassettes Ready for Vector Ligation and Yeast Transformation
At this point, Golden Gate Assembly compatible BsaI sites have been added to the 5 necessary violacein biosythesis expression cassettes via overhang PCR. The Herculase Fusion II polymerase was used in the reaction to lower the probability of point mutations. The following Gel shows each expression cassette digested with BsaI prior to gel extraction and purification.
Violacein loxPsym Integration
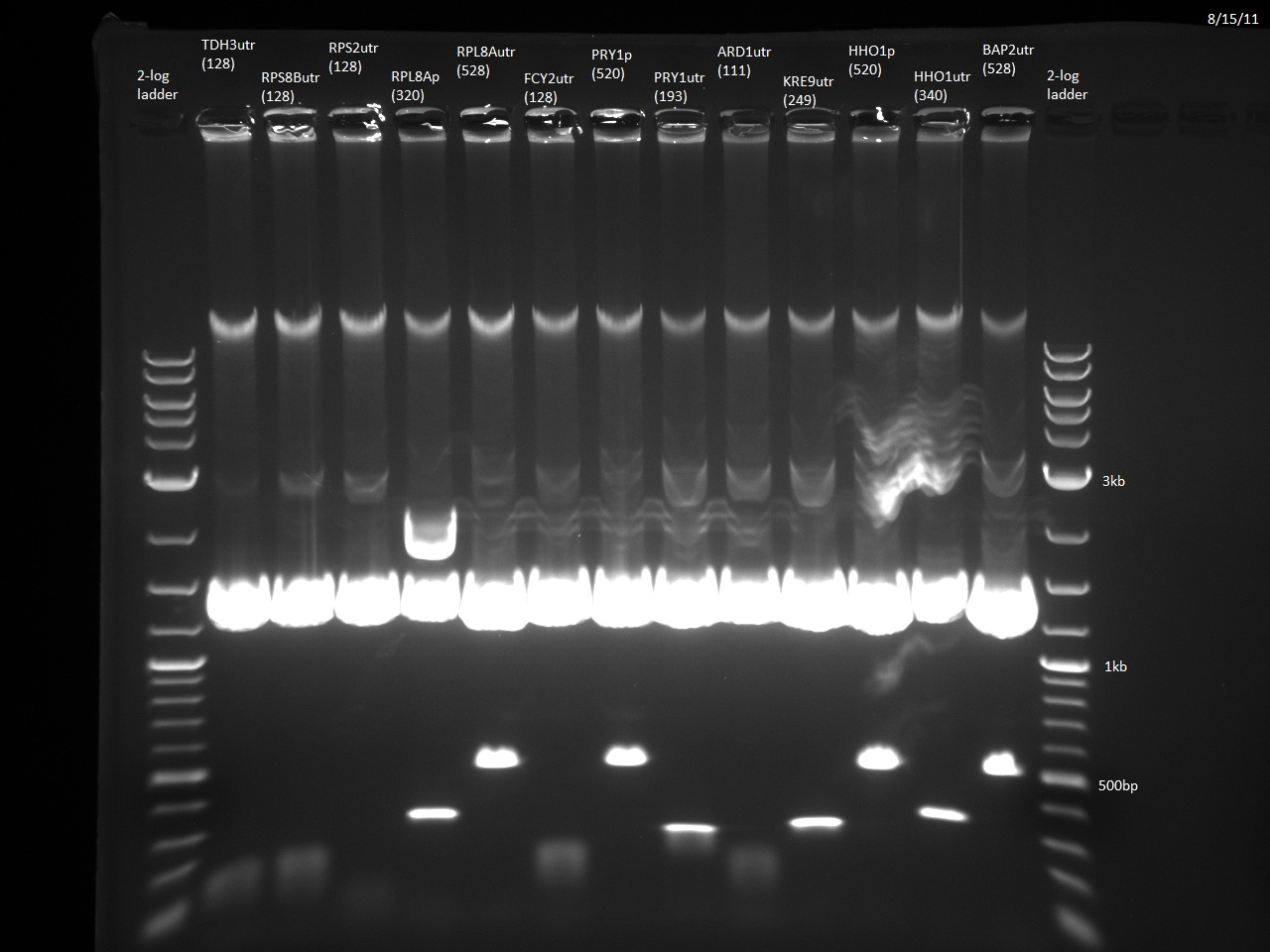
We currently have 23/24 promoters and 3'UTRs from our Golden Gate Assembly-compatible library blunt-end ligated into the vector pTZ19u with SmaI. The promoters and 3'UTRs were PCR'd from yeast genomic DNA with primers containing designed BsaI overhangs. Following miniprep and digestion with BsaI, the promoters and 3'UTRs can be combined with our violacein ORFs. The last fragment, RPS2utr, has had the BsaI overhangs added, and will soon be ligated into the vector and transformed into E. coli. Additionally, the TDH3 3'UTR has been ligated to the URA3 sequence using the NotI site, which will allow the selection of yeast that has taken up our 'expression circles'. Finally, the loxPsym linker has been constructed from two single strand DNA oligos. This means that we are almost ready to do the random ligation to create the combinatorial library of violacein 'expression circles' and integrate them into yeast using Cre-Lox recombination.
 "
"