Team:Groningen/project characterisation mu
From 2011.igem.org
| Line 85: | Line 85: | ||
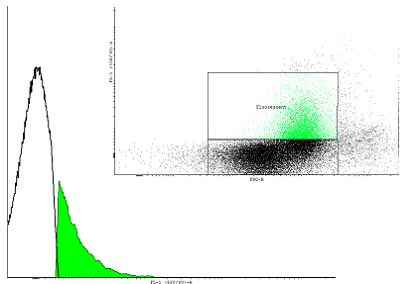
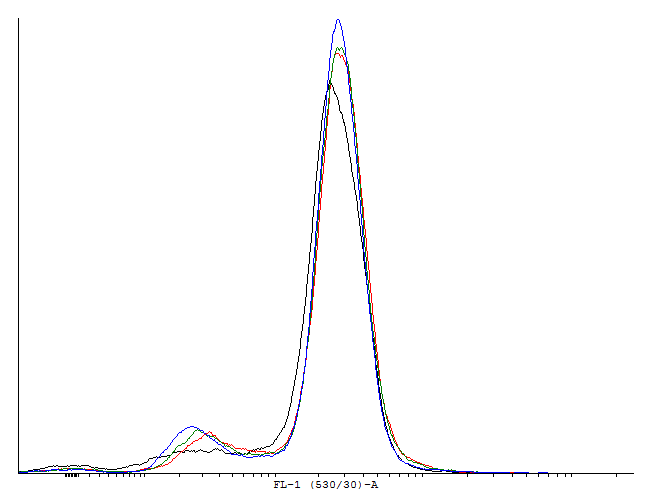
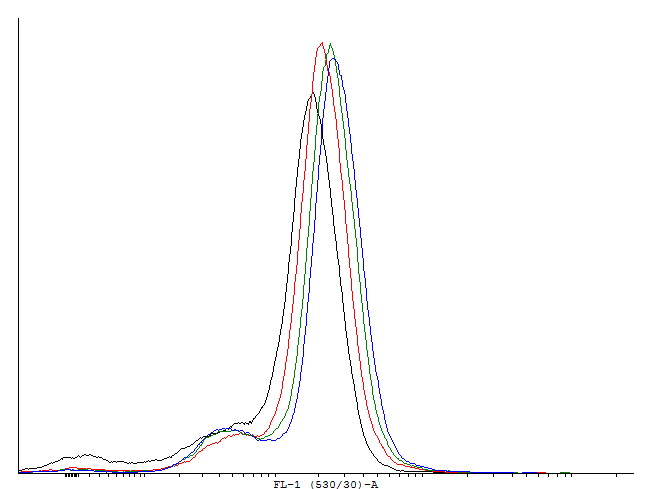
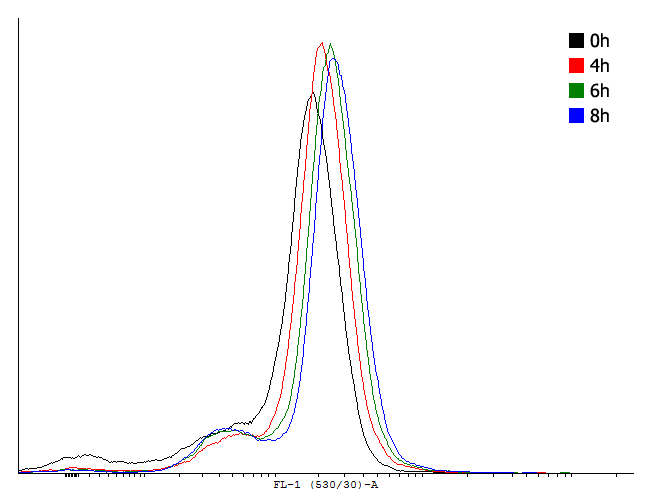
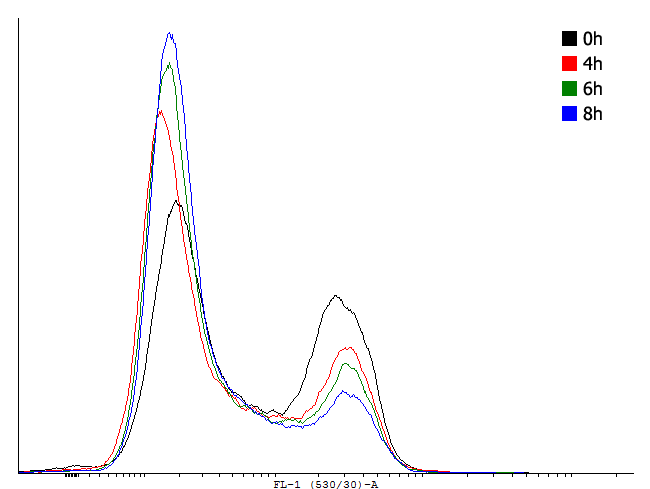
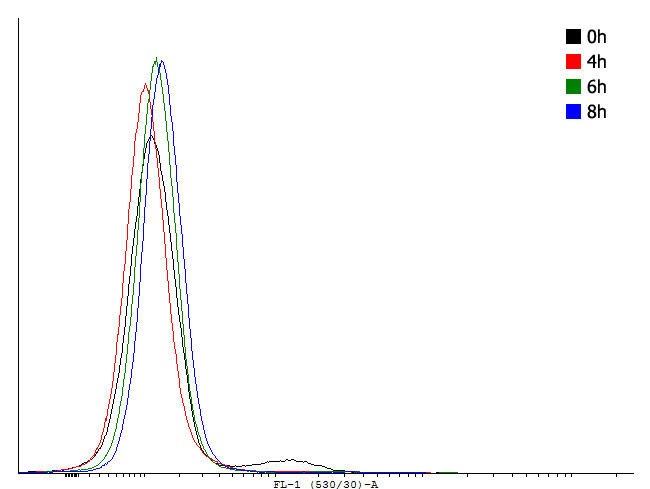
Figure 1. This autoinducing loop does not exhibit heterogenity when non-induced. Only the population that is not producing GFP is observed. That means that the loop is not induced through a leaky promoter and is a good candidate for an inducible bistable switch. | Figure 1. This autoinducing loop does not exhibit heterogenity when non-induced. Only the population that is not producing GFP is observed. That means that the loop is not induced through a leaky promoter and is a good candidate for an inducible bistable switch. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{{FooterGroningen2011}} | {{FooterGroningen2011}} | ||
Revision as of 17:12, 21 September 2011
Memory Units
cI Autoinducing Loops
The autoinducing loop driven by cI will function as our first memory unit. We aimed to find a combination of RBS, transcription factor and degradation tag that enables us to toggle between the two steady states in a robust way. To estimate the rate in which single cells are activated by too much leakage of cI protein we measured the fluorescence in uninduced cells with a flow cytometer. To measure the activation we used a construct with the PRM promoter producing GFP with an LVA tag to make dynamic measurement possible.
| cI autoinducing loop 6 (BBa_K00) after 4 hours of incubation | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
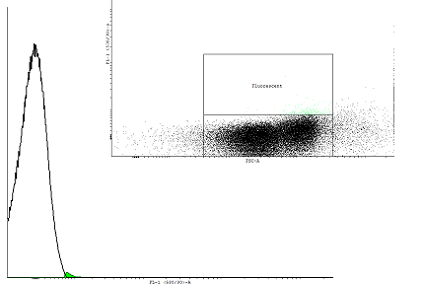
Figure 2. A dotplot of uninduced cells carrying the pSB1K3 plasmid with the biobrick BBa_ and the plasmid pSB1A3 with as well an input(PBAD controlling cI) as well as the output (PRM controlling GFP-LVA). |
| ||||||||||||||||
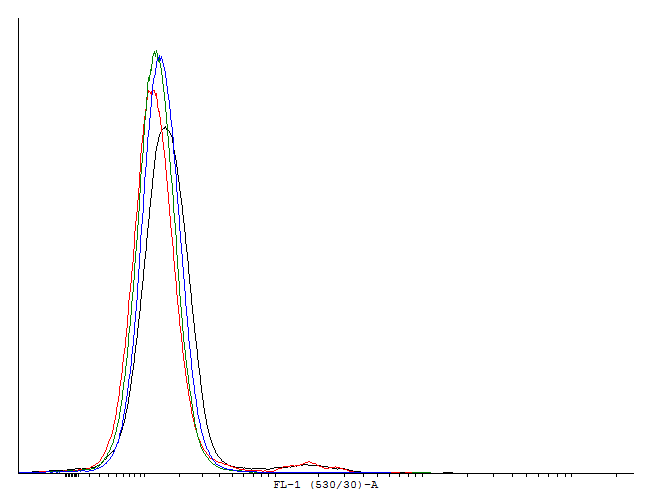
| Only one population is visible on the dotplot. The histogram was obtained in the same fashion as in figure 1 to show that there are cultures with activated cI loops and cultures without. | |||||||||||||||||
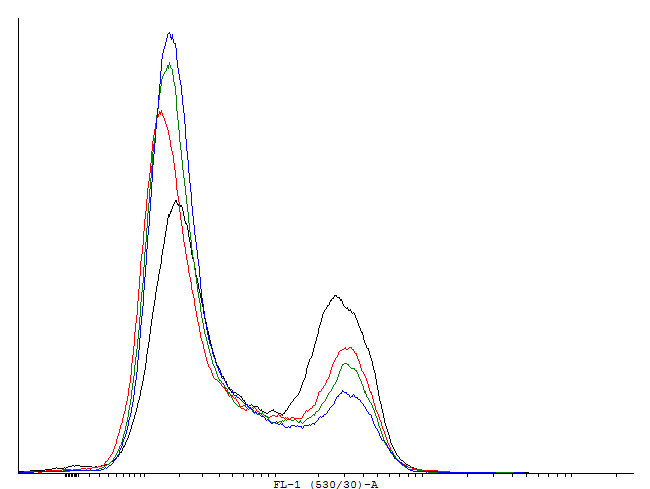
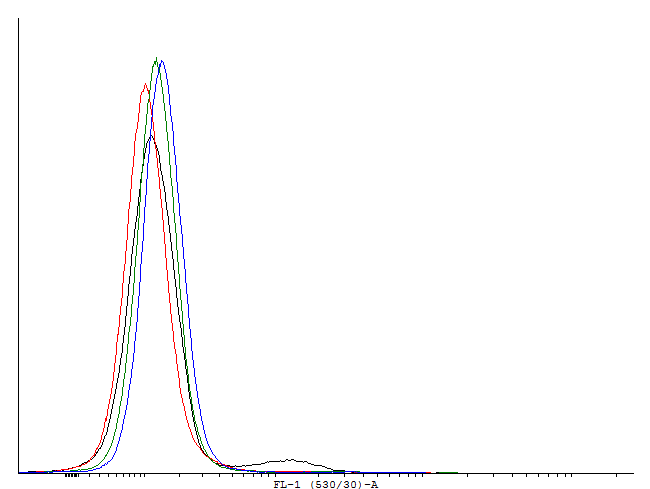
These measurements indicate activation and by this bimodal behaviour of cells containig BBa_. But no activation of BBa_ and several other constructs. In these measurements we used GFP-LVA. The degradation seems to be about as fast as the production so only faint signals are visible. We decided to repeat the measurement with similar constructs but with GFP instead of GFP-LVA as an output.
| no-tag | DAS | LVA | |||||
| RBS 0,07 | |
| | 
| |||
| RBS 0,3 | | | | ||||
| RBS 0,6 |
| | | ||||
| RBS 1 | | | | ||||
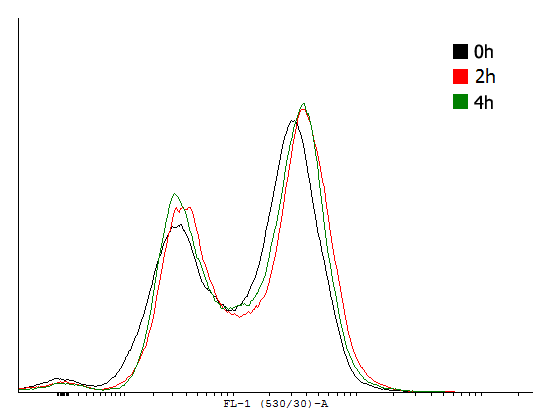
| Flow cytometer data taken at different time-points during exponential growth of cells carrying different cI autoinducing loops. | |||||||
This result shows that both, tag and RBS, used for the expression of the transcription factor have a huge impact on the bimodal behavior of the autoinducing loop. All loops that contain a LVA-tag tag are inactive if not induced. Those without any tag are mostly active even without induction while those with a DAS-tag show a bimodal behavior. Especially the cells carrying the plasmids with a strong RBS in the autoinducing loop look promising.
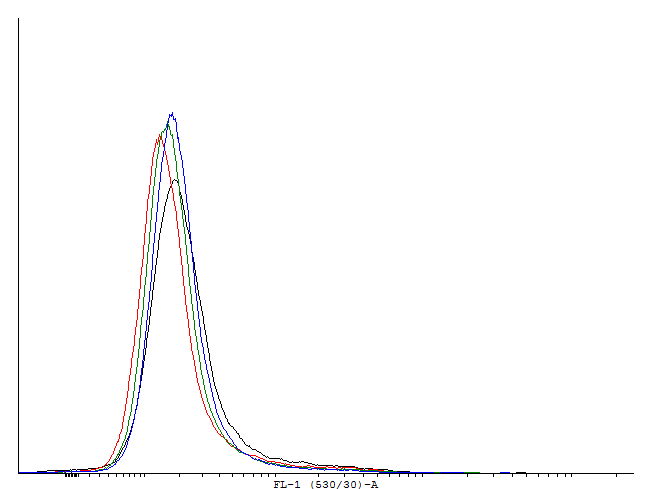
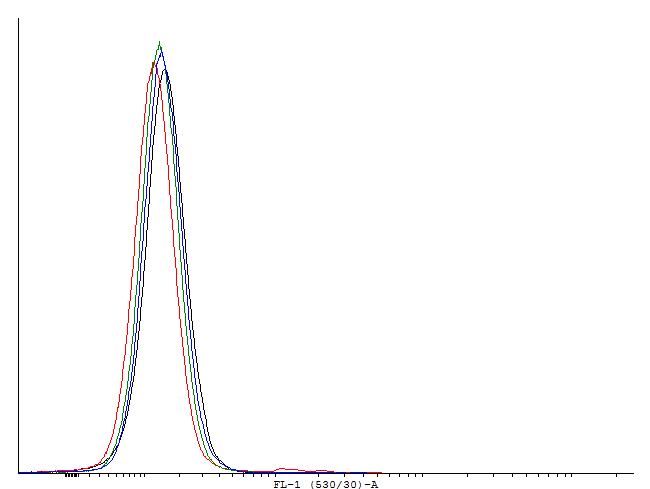
This loop is clearly activated.
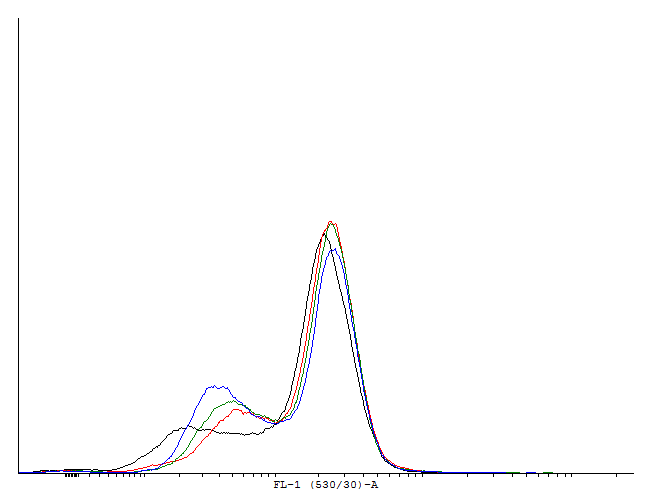
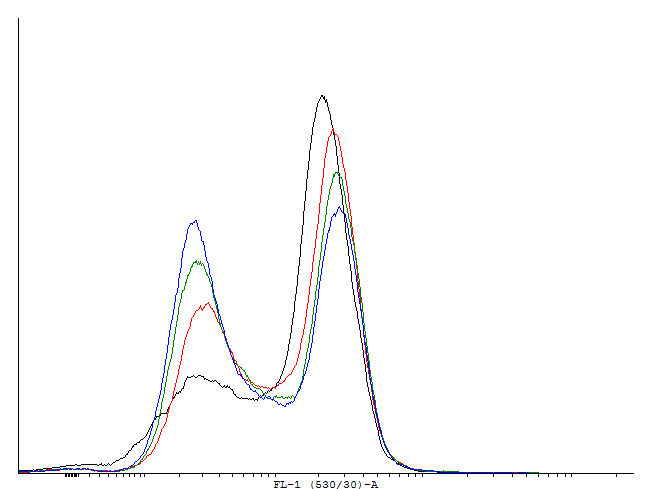
This loop is bimodal.
Figure 1. This autoinducing loop does not exhibit heterogenity when non-induced. Only the population that is not producing GFP is observed. That means that the loop is not induced through a leaky promoter and is a good candidate for an inducible bistable switch.
 "
"