Team:Potsdam Bioware/Labjournal/July
From 2011.igem.org
28th Labday 2011-07-01
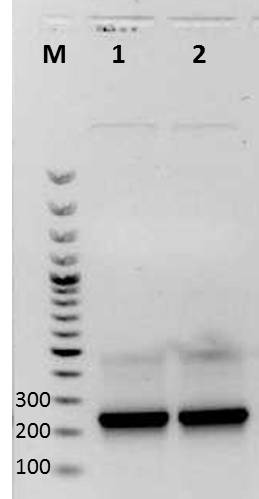
Agarose gel electrophoresis of PCR (mdnA for phage display, strategy 1)
Investigators: Sabine, Sandrina
Aim: Verification of mdnA for phage display (strategy 1)
Time: 2011-06-30,14:00-15:30
Materials:
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- PCR products: mdnA for phage display (strategy 1) from 2011-06-30
- DNA Ladder Gene Ruler, 100bp plus (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of a 1 % agarose gel
- mAgarose = 0.5 g in 50 ml 1x TAE
- adding 2 µl gel red
2. Loading samples
- 5 µl PCR product and 6x loading dye
- 2 µl DNA ladder gene ruler (1:10)
3. Run
- 100 V
- time: 00:45 h
Results:
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | marker | 2 | |
| 1 | mdnA with SfiI | 6 | 224 |
further tasks:
purification of PCR product
purification of PCR product mdnA SfiI for phage display
Investigators: Sabine, Sandrina
Aim:
- purification of PCR product mdnA with SfiI restriction sites for phage display (strategy 1)
Time: 2011-06-24, 15:30-16:00
Materials/Methods:
- QIAquick Gel Extraction Kit (250)
Further tasks:
- digestion of the fragments with SfiI
Agarose gel electrophoresis of myc-tagged mdnA
Investigators: Jessica
Aim: Verification of myc-tagged mdnA
Time: 2011-07-01,13:00-15:30
Materials:
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- PCR product: myc-tagged mdnA from 2011-06-29
- DNA Ladder Gene Ruler, 100bp plus (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of a 1 % agarose gel
- mAgarose = 0.5 g in 50 ml 1x TAE
- adding 2 µl gel red
2. Loading samples
- 45 µl PCR product and 9 µl 6x loading dye
- 6 µl DNA Ladder Gene Ruler (1:10)
3. Run
- 100 V
- time: 1 h
Results:
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | marker | 6 | |
| 1 | myc-tagged mdnA | 27 | 233 |
| 2 | myc-tagged mdnA | 27 | 233 |
- Gel excision of bands, stored at -20 °C (PCR box, blue)
Further tasks:
- Gel extraction
- Restriction digest
29th Labday 2011-07-04
digestion of PCR product mdnA with restriction enzyme SfiI (strategy 1)
Investigators: Sandrina, Sabine
Aim: digestion of mdnA with SfiI for cloning into pak100
Time: 2011-07-04,14:00-16:30
Materials:
Digestion with SfiI
- 50 µl sample
- NEB 10x buffer 4
- 100x BSA
- 3 µl restriction enzyme SfiI
- PCR product (mdnA with SfiI restriction sites)
Further tasks:
ligation with pak100bla-KDIR
ligation of mdnA into pak100(strategy 1)
Investigators: Sandrina, Sabine
Aim: ligation of mdnA into pak100 for phage display
Time: 2011-07-04,16:00-16:30
Materials:
Ligation: 2 µl vector (pak100 bla KDIR, digested with SfiI), 2 µl insert (mdnA, digested with SfiI), 1 µl quick ligase, 5 µl 2x quick ligase NEB, 10 min, room temperature
Further tasks:
purification
Realization:
amplified mdnA does not contain a myc-tag --> further task: order a new reversed primer for mdnA (strategy 1) containing a myc-tag
Amplification of mdnA for phage display (strategy 2)
Investigators: Sandrina, Sabine
Aim: amplificate mdnA of pARW089 with primers pf_mdnA_sfi_1 and pr_mdnA_sfi_1
Time: 2011-06-10,14:00-16:30
Materials/Methods:
PCR
- 5 µl Vector pARW089
- Eppendorf Mastercycler Gradient (program sabrina)
- 0,25 µl OneTaq DNA Polymerase (NEB)
- 1 µl dNTPs
- 1 µl per primer
- 31,75 µl DNase free water
- 10 µl 5xOneTaq Standard Reaction Buffer
Gel electrophoresis PCR product mdnA (strategy 2)
Investigators: Sandrina, Sabine
Aim:verification of mdnA (strategy 2)
Time: 2011-07-04,17:00-19:00
Materials/Methods:
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- PCR products: mdnA for phage display (strategy 1
- DNA Ladder Gene Ruler, 100bp plus (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of a 1 % agarose gel
- mAgarose = 0.5 g in 50 ml 1x TAE
- adding 2 µl gel red
2. Loading samples
- 5 µl PCR product and 6x loading dye
- 2 µl DNA ladder gene ruler (1:10)
3. Run
- 100 V
- time: 01:15 h
Results:
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| 1 | marker | ||
| 2 | PCR product mdnA | 5 | ca. 230 |
30th Labday 2011-07-06
digestion of PCR products mdnA, geneIII and vector pARW089 (strategy 2)
Investigators: Sandrina, Sabine
Aim: digestion of mdnA, gene III and pARW089 for getting an mdnA-geneIII fusion gene with rfc25 restrition sites in pARW089 by ligation of the three fragments
Time: 2011-07-06,14:00-16:00
Materials:
Digestion of mdnA with NarI (EheI isoschizomere) and AgeI
- 40 µl sample
- NEB 10x buffer (4 µl)
- 1 µl restriction enzyme NarI
- 1 µ restriction enzyme AgeI
- 30 µl PCR product (mdnA with rcf 25 and EheI/NarI restriction sites)
- 4 µl H²O
Digestion of geneIII with NgoMIV and AatII
- 40 µl sample
- NEB 10x buffer (4 µl)
- 1 µl restriction enzyme NgoMIV
- 1 µ restriction enzyme AatII
- 30 µl PCR product (geneIII with rcf 25 and AatII restriction sites)
- 4 µl H²O
Digestion of vector pARW089 with NarI (EheI isoschizomere) and AatII
- 20 µl sample
- NEB 10x buffer (2 µl)
- 1 µl restriction enzyme NarI
- 1 µ restriction enzyme AatII
- 12 µl vector DNA
- 4 µl H²O
- 1 h at 37°C
Further Tasks:
- gel electrophoresis and purification of the three digested fragments
Gel extraction from agarose gel of digested fragments mdnA, geneIII and pARW089 (strategy 2)
Investigators: Stefan
Aim: Purification of mdnA, geneIII and pARW089 (strategy 2) to obtain an mdnA-geneIII fusion gene which can be cloned into pARW089
Time: 2011-07-06,17:00-19:00
Materials/Methods:
- PCR Clean-up Kit NucleoSpin ExtractII (Machery-Nagel)
Method:
1. Cut out visible DNA bands of the digestion product of a 0.8% and 1% agarose gel, respectivly
2. Use protocol of PCR clean-up
3. Two bands were cut out for the vector and genIII. Just use the lower two bands for first ligation.
Results:
- DNA concentration geneIII: 5,4 ng/µl
- DNA concentration mdnA: 8,2 ng/µl
- DNA concentration pARW089: 3,8 ng/µl
- measured by NanoDrop
Further Tasks:
- Repeat digestion of pARW089 (because of low concentration)and ligation of the three fragments
31th Labday 2011-07-07
repeated digestion of pARW089 with NarI and AatII (strategy 2)
Investigators: Sabine, Sandrina
Aim: Digestion of pARW089 (strategy 2) so that an mdnA-geneIII fusion gene can be cloned into pARW089
Time: 2011-07-07,10:00-12:00
Materials/Methods:
Digestion of vector pARW089 with NarI (EheI isoschizomere) and AatII
- 20 µl sample
- NEB 10x buffer (2 µl)
- 1 µl restriction enzyme NarI
- 1 µ restriction enzyme AatII
- 3 µl vector DNA (ca. 800 ng/ µl)
- 13 µl H²O
- 2 1/2 h at 37°C
- afterwards: inactivation of restriction enzymes: 20 min, 65°C
Further tasks:
gel electrophoresis of digested vector
Gel electrophoresis of digested pARW089 vector(strategy 2)
Investigators: Sandrina, Sabine
Aim:Purification of pARW089 (strategy 2) so that an mdnA-geneIII fusion gene can be cloned into pARW089
Time: 2011-07-07,14:30-15:30
Materials/Methods:
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- digested vector pARW089
- DNA Ladder Gene Ruler 1kb plus(Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of a 0,5 % agarose gel
- mAgarose = 0.25 g in 50 ml 1x TAE
- adding 2 µl gel red
2. Loading samples
- digestion product and 6x loading dye
- pARW089 digestion product (first try, conc: 3,8 ng/µl) to verify measured concentrations
3. Run
- 100 V
- time: 00:45 h
Results:
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| 1 | marker | ||
| 2 | purificated, digested vetor try 1 | 30 | ca. 10 000 |
| 3 | digested pARW089 vector | 30 | ca. 10.000 |
File:UP versuch2 pARW Verdau.7.7.png
one band was cutted out of the gel for purifacation
Further tasks:
ligation of mdnA, geneIII and pARW089
ligation of mdnA and geneIII into pARW089(strategy 2)
Investigators: Sabine
Aim: ligation of mdnA, geneIII, and pARW089 to get an mdnA-geneIII fusion gene in pARW089for phage display
Time: 2011-07-07,15:30-17:30
Materials:
Ligation: 2 µl vector (pARW089 digested with NarI and AatII), 3 µl insert (mdnA digested with NarI and AgeI, and geneIII digested with AatII and NgoMIV), 1 µl T4 ligase, 1 µl T4 ligase NEB-buffer,over night, 16°C
Further tasks:
transformation of E. coli cells(XL1-blue)
32th Labday 2011-07-12
Transformation of ligation mdnA, geneIII, and pARW089 (2011-07-07) for phage display (strategy 2)
Investigators: Sandrina, Sabine
Aim:amplification of previous ligation
Time: 9:00-11:30
Method:
- ligation-samples from 2011-07-07;
(ligation of mdnA, geneIII and pARW089)
protocol:
addition of 10 µl ligation reaction to cells (XL1-blue, tet-resistance) in 1.5 ml Eppi,
incubation 25 min on ice,
heat shock 45 sec at 42°C,
incubation 2 min on ice,
addition of 750 µl LB medium,
incubation at 37 °C for 60 min in Eppendorf thermomixer at 750 rpm,
plating on LB medium with 1,5 % agar, 100 µg/ml ampicillin, 100 µg/µl tetracyclin
storage over night at 37°C
Further tasks:
control cell clones
33th Labday 2011-07-13
Error prone PCR (varied MnCl2 concentration) of mdnA
Investigators: Steffi, Jessica, Katharina
Time: 2011-06-29,14:00-16:00
Aim: Random mutation of mdnA gene based on increased MnCl2 concentration
Materials and Methods: see 26th Labday
34th Labday 2011-07-14
Agarose gel electrophoresis of error prone PCR of mdnA from 2011-07-13 and gel extraction
Investigators: Katharina
Time: 2011-06-29,8:00-11:00
Materials:
- Agarose broad range (Roth)
- 1x TAE buffer Gel Red
- 10 PCR products of mutated mdnA
- DNA Ladder GeneRuler, 100bp plus (1:10) (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of two 1 % agarose gel
For each gel: mAgarose = 0.5 g in 50 ml 1x TAE adding 2 µl gel red
2. Loading samples
50 µl PCR product and 6x loading dye
6 µl DNA Ladder GeneRuler (1:10) (Fermentas)
3. Run
- 110 V time: 1 h
Results:
| gel 1 | gel 2 | ||||||
|---|---|---|---|---|---|---|---|
| lane | Sample | Volume in µl | Expected size in bp | Sample | Volume in µl | Expected size in bp | |
| 1 | marker | 6 | marker | 6 | |||
| 2 | PCR sample 1a | 50 | 276 | PCR sample 1b | 50 | 276 | |
| 3 | PCR sample 2a | 50 | 276 | PCR sample 2b | 50 | 276 | |
| 4 | PCR sample 3a | 50 | 276 | PCR sample 3b | 50 | 276 | |
| 5 | PCR sample 4a | 50 | 276 | PCR sample 4b | 36 | 276 | |
| 6 | PCR sample 5a | 50 | 276 | PCR sample 5b | 50 | 276 | |
The bands were excised and purified using the NucleoSpin Extract II (Macherey-Nagel) extraction Kit.
Further Tasks:
- digestion of the purified samples
- ligation
Digestion of error prone PCR of mdnA from 2011-07-13 and pARW089 and agarose gel electrophoresis
Investigators: Jessica
Time: 2011-06-29,11:00-15:00
Materials:
- Restriction enzymes: AatII, NarI
- NEB Buffer 4
- PCR products of random mutated mdnA(1a-5a), purified from gel
- vector pARW089
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- GeneRuler DNA Ladder Mix (1:10) (Fermentas)
- GeneRuler 1kb DNA Ladder (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Digestion:
| components | volume of PCR products /µl | volume of pARW089 /µl |
| DNA | 30 | 10 |
| NEB Buffer 4(10x) | 4 | 4 |
| Enzyme AatII | 0.5 | 0.8 |
| Enzyme NarI | 2 | 2 |
| H2O | 3.5 | 23.2 |
| Total volume | 40 | 40 |
- digestion at 37°C for 1 h
2. Production of agarose gel
- 1.7% gel for PCR products: mAgarose = 0.85 g in 50 ml 1x TAE adding 2 µl gel red
- 0.8% gel for vector: mAgarose = 0.4 g in 50 ml 1x TAE adding 2 µl gel red
3. Loading samples
- 40 µl digest and 8 µl 6x loading dye
4. Run
- 80 V, time: >1 h
Results:
Gel 1
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | GeneRuler DNA Ladder Mix (1:10) | 6 | |
| 1 | digested mdnA 1a | 48 | |
| 2 | digested mdnA 2a | 48 | |
| 3 | digested mdnA 3a | 48 | |
| 4 | digested mdnA 4a | 48 | |
| 5 | digested mdnA 5a | 48 |
Gel 2
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | GeneRuler 1kb DNA Ladder | 2 | |
| 1 | digested pARW089 | 48 |
- Gel excision and storage at -20°C (DNA box, orange)
Overnight culture of PDV089
Investigators: Sandrina, Sabine
Aim:
control plasmid ligation (pARW089, mdnA, geneIII --> PDV089, strategy 2)
Time: 2011-07-13,12:00-13:00
Materials/Methods:
- LB-medium with tet and amp
- cell clones from over night plate
- incubate over night at 37°C and 750 rpm
Further tasks:
- plasmid preparation and analytic digestion
Repeated PCR of mdnA for phage display (strategy 1) repeated with new ordered reversed primer (with myc-tag) and geneIII
Investigators: Sandrina, Sabine
Time: 2011-06-30,13:00-16:00
Aim:
- amplification of mdnA with SfiI restriction sites and myc (vector pARW089) to clone it into pAk100 bla KDIR
- amplificate GeneIII of pak100 with primers pf_iGEM_GenIII_NgoMIV and pr_GenIII_iGEM_AatII (phage display strategy 2), because there were two bands after digestion with NgoMIV and AatII after gel electrophoresis
Materials/Methods:
see 2011-06-10
changes:
- program: 123, Thermo Hybrid, PX2
further tasks:
gel electrophoresis with PCR product
35th Labday 2011-07-15
Ligation of mutated mdnA into pARW089
Investigators: Katharina
Aim: create pARW089 vector with inserted mutated mdnA
Time: 10:00-14:00
Materials:
Ligation: 2 µl vector (pARW089 digested with NarI and AatII), 3 µl insert (mdnA digested with NarI and AatII), 1 µl T4 ligase, 1 µl T4 ligase NEB-buffer, 4 h at room temperature
Further tasks:
transformation of E. coli cells(XL1-blue)
Transformation of competent E. coli cells with mutated mdnA in pARW089
Investigators: Katharina
Aim:Transformation of ligations
Time: 14:00-15:00
Method:
protocol:
addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
incubation 25 min on ice,
heat shock 45 sec at 42°C,
incubation 5 min on ice,
addition of 750 µl LB medium,
incubation at 37 °C for 60 min in Eppendorf thermomixer at 750 rpm,
plating on LB medium with 1,5 % agar, 100 µg/ml kanamycin
storage over night at 37°C
Further tasks:
control cell clones
ligation of mdnA into pak100(strategy 1)
Investigators: Sandrina, Sabine
Aim: ligation of mdnA into pak100 for phage display
Time: 2011-07-15,11:00-13:00
Materials:
Ligation: 2 µl vector (pak100 bla KDIR, digested with SfiI), 2 µl insert (mdnA, digested with SfiI), 1 µl T4 ligase, 5 µl 2x 10x T4 ligase buffer, 20 min, room temperature
Further tasks:
transformation into competent cells
Transformation of ligation mdnA into PAK100 bla KDIR for phage display (strategy 1)
Investigators: Sandrina, Sabine
Aim:amplification of previous ligation
Time: 13.00-15:00
Method:
- ligation-samples
(ligation of mdnA into PAK100 bla KDIR)
protocol:
addition of 10 µl ligation reaction to cells (XL1-blue, tet-resistance) in 1.5 ml Eppi,
incubation 25 min on ice,
heat shock 45 sec at 42°C,
incubation 2 min on ice,
addition of 750 µl LB medium,
incubation at 37 °C for 60 min in Eppendorf thermomixer at 750 rpm,
plating on LB medium with 1,5 % agar, 100 µg/ml ampicillin, 100 µg/µl tetracyclin
storage over night at 37°C
Further tasks:
control cell clones
ligation of mdnA and geneIII for strategy 2
Investigators: Sabine, Sandrina
Aim:ligation of mdnA and geneIII to get an mdnA-geneIII fusion gene phage display
Time: 2011-07-15,15:00-16:00
Materials:
Ligation: 3 µl insert mdnA digested with NarI and AgeI and geneIII digested with AatII and NgoMIV), 1 µl T4 ligase, 1 µl T4 ligase NEB-buffer, 2 µl H2O , 20 min, room temperature
Results:
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | marker, DNA ladder mix ladder mix | 10 | |
| 1 | mdnA ligated with geneIII | ca. 600 |
Further tasks:
ligation with pARW089
ligation of the fusion gene mdnA-geneIII into pARW089(strategy 2)
Investigators: Sabine, Sandrina
Aim:ligation of mdnA-geneIII in pARW089 to get an mdnA-geneIII fusion gene in pARW089for phage display
Time: 2011-07-07,17:00-19:00
Materials:
- Ligation: 2 µl vector (pARW089 digested with NarI and AatII), 3 µl insert, 1 µl T4 ligase, 1 µl T4 ligase NEB-buffer,over night, 16°C
- two bands were observed after mdnA-geneIII ligation (red boxes)--> ligation with pARW089 was tried with both bands
Further tasks:
transformation of E. coli cells(XL1-blue)
36th Labday 2011-07-16
Transformation of ligated pARW089 with mdnA-geneIII
Investigators: Stefan
Aim:Transformation of phage display ligations (pARW089 with mdnA-geneIII) using heat shock
Time: 12:00-20:00
Method/Materials:
1) Take chemically competent E. coli XL1 blu cells from –80°C freezer.
2) Turn on heat block to 42°C.
3) use 60 µL of competent cells for each approach
4) Keep tubes on ice.
5) Add 2 µL DNA solution into the E. coli cell suspension, mix by flicking the tube. Incubate on ice for 15-30 min.
6) Put tubes into heat block at 42°C for 45 seconds.
7) Put tubes back on ice for 2 minutes to reduce damage to the E. coli cells.
8) Add 750 µL of LB with no antibiotic. Incubate tubes for 1 hour at 37°C and 750 rpm.
9) spin down the cell suspension for 3 min at 6000 rcf at 4 °C , discard most of the supernatant, resuspend the bacterial pellet by pipetting and use this for spreading.
10) Spread 100 ul of the resulting culture on LB agar plates with Tet/CM.
11) Grow overnight at 37 °C.
12) Pick colonies about 12-16 hours later.
Further tasks:
picking clones and plasmid preparation
Overnight culture of XL1 blue with mdnA in PAK100blaKDIR
Investigators: Stefan
Aim: Picking clone 1 from plate and do an overnight culture
Time: 12:00-20:00
Method/Materials:
- Clone ID_1: Strain:XL1 blue, Plasmid:mdnA in PAR100bla KDIR, Clone number:1
15 mL of LB media were supplied with 15 µL of Tet and CM and cells were incubated at 37 °C overnight.
Further tasks:
plasmid preparation
37th Labday 2011-07-17
Miniprep of E. coli overnight culture containing PAK100blaKDIR
Investigators: Stefan
Aim: isolate the plasmid
Time: 12:00-20:00
Method/Materials:
protocol 5.1 of the NucleoSpin Plasmid Kit was used
Further tasks:
sequence the plasmid
E. coli XL1 blue stock culture containing PAK100blaKDIR with mdnA (Phage Display-strategy 1)
Investigators: Stefan
Aim: set up a stock culture
Time: 12:00-20:00
Method/Materials:
2 x 700 µL culture were mixed with 300 µL of glycerol and stored in -80 °C freezer
Further tasks:
overnight culture of E. coli Xl blue m. Ligation089jessi, u.Ligationvector089jessi,u.Ligationu.BandemdnAgeneIIIpARW089, mLigatiojnm.BandemdnAgeneIIIpARW089
Investigators: Stefan
Aim: pick two clones each and inoculate them overnight in 15 mL LB supplied with 15 µL Tet and Amp
Time: 12:00-20:00
Method/Materials:
Further tasks:
plasmid preparation
38th Labday 2011-07-18
overnight culture of E. coli XL-1 blue containing mutated mndA in parW089
Investigators: Katharina
Aim: Picking 5 clones from each plate and do an overnight culture
Time: 14:00-15:00
Method/Materials:
E. coli Strain:XL1 blue, Plasmid:mdnA in parW089
5 mL of LB media were supplied with 5 µL of Kan and cells were incubated at 37 °C overnight.
Further tasks:
plasmid preparation
Dual-Tranformation of XL1-blu (E. coli) with pTEV-SCS1 and pJC354_ssTorA_NheI_CS-TEV_XhoI_bla
Investigators: Sascha
Aim:
- Dual-Transformation of XL1-blue to test the in-vivo selection system (pTEV-SCS1 + pJC354_ssTorA_NheI_CS-TEV_XhoI_bla)
Materials:
- Plasmids:
- pTEV-SCS1 (TEV protease, res. against kanamycin)
- pJC354_ssTorA_NheI_CS-TEV_XhoI_bla (res. against chloramphenicol)
- pJC354_ssTorA_NheI_CS-TEV_XhoI_bla (res. against chloramphenicol)
- competent cells XL1-blue
- LB media and diffenet antibiotics
Used method:
- standard protocol for heat shock transformation
Output: transformed cultures with plasmids
- XL1-blue without plasmid (negativ control - no growth over night)
- XL1-blue transformed with pTEV-SCS1 - growth with kanamycin in LB media
- XL1-blue transformed with pJC354_ssTorA_NheI_CS-TEV_XhoI_bla - growth with chloramphenicol in LB media
- XL1-blue transformed with pTEV-SCS1 and pJC354_ssTorA_NheI_CS-TEV_XhoI_bla - growth with kanamycin and chloramphenicol in LB media
Further tasks:
- reinfect media with cultures for new growth period and distribution to LB Agar with different antibiotic concentrations
Test digestion of ligations for strategies 1 and 2
Investigators: Sandrina
Aim:control if liagation of geneIII and mdnA into pARW089 (strategy 2) and ligation of mdnA into PAK100 bla KDIR worked
Time: 15:30-19:00
Materials/Methods:
Strategy 1:
- 0,5 µl SfiI
- 2 µl 10x buffer 4 (NEB)
- 0,2 µl BSA
- 10 µl vector DNA
- 13,3 µl H2O
incubate for 1 h at 50°C
Strategy 2:
- 0,5 µl XbaI
- 0,5 µl SpeI
- 2 µl 10x buffer 4 (NEB)
- 0,2 µl BSA
- 10 µl vector DNA
- 12,8 µl H2O
incubate for 1 h at 37°C
Results:
- expected bands: strategy 1: ca. 5000 bp and 200bp
- expected bands: strategy 2: ca. 5000 bp, 4000 bp, 600 bp and 200
observed bands:
- srtategy 1: ca. 5000 and 800 bp
- strategy 2: ca. 5000 and 200 bp
Further tasks:
- repeat ligations
39th Labday 2011-07-19
Resistance test of the Dual-Transformation of XL1-blue (E. coli) with pTEV-SCS1 and pJC354_ssTorA_NheI_CS-TEV_XhoI_bla
Investigators: Stefan and Sebastian
Aim:
- Dual-Transformation of XL1-blue to test the in-vivo selection system (pTEV-SCS1 + pJC354_ssTorA_NheI_CS-TEV_XhoI_bla)
Materials:
- Overnight cultures
- XL1-blue transformed with pTEV-SCS1 - growth with kanamycin in LB media
- XL1-blue transformed with pJC354_ssTorA_NheI_CS-TEV_XhoI_bla - growth with chloramphenicol in LB media
- XL1-blue transformed with pTEV-SCS1 and pJC354_ssTorA_NheI_CS-TEV_XhoI_bla - growth with kanamycin and chloramphenicol in LB media
- LB media and different antibiotics
Used method:
- growth over night
- set OD (600 nm) after measurement cells were dilutes to OD 0,1, transformed XL1-blue grow again to OD 0,5. Out of this approach a dilution to OD= 0.002 was done and platted to LB agar with kanamycin, IPTG and different concentrations of ampicillin (range form 0-800 ng/ml ampicillin)
Output:
- survival of dual transformed cells...
Further tasks:
There was no growth on the TorAbla plates due to the choice of the wrong antibiotic. This was set up again as well as well as the double transformed cells.
Stock cultures of XL1-blue(E. coli)with pTEV-SCS1 and pJC354_ssTorA_NheI_CS-TEV_XhoI_bla and of XL1-blue(E. coli)with pTEV-SCS1 and of XL1-blue(E. coli)pJC354_ssTorA_NheI_CS-TEV_XhoI_bla
Investigators: Stefan
Aim:
- make stock cultures
Materials:
- Overnight cultures were used. 700 µL culture were spun down to remove antibiotics. The pellets is resuspended in 700 µL LB media and 300 µL glycerol.
Used method:
Further tasks:
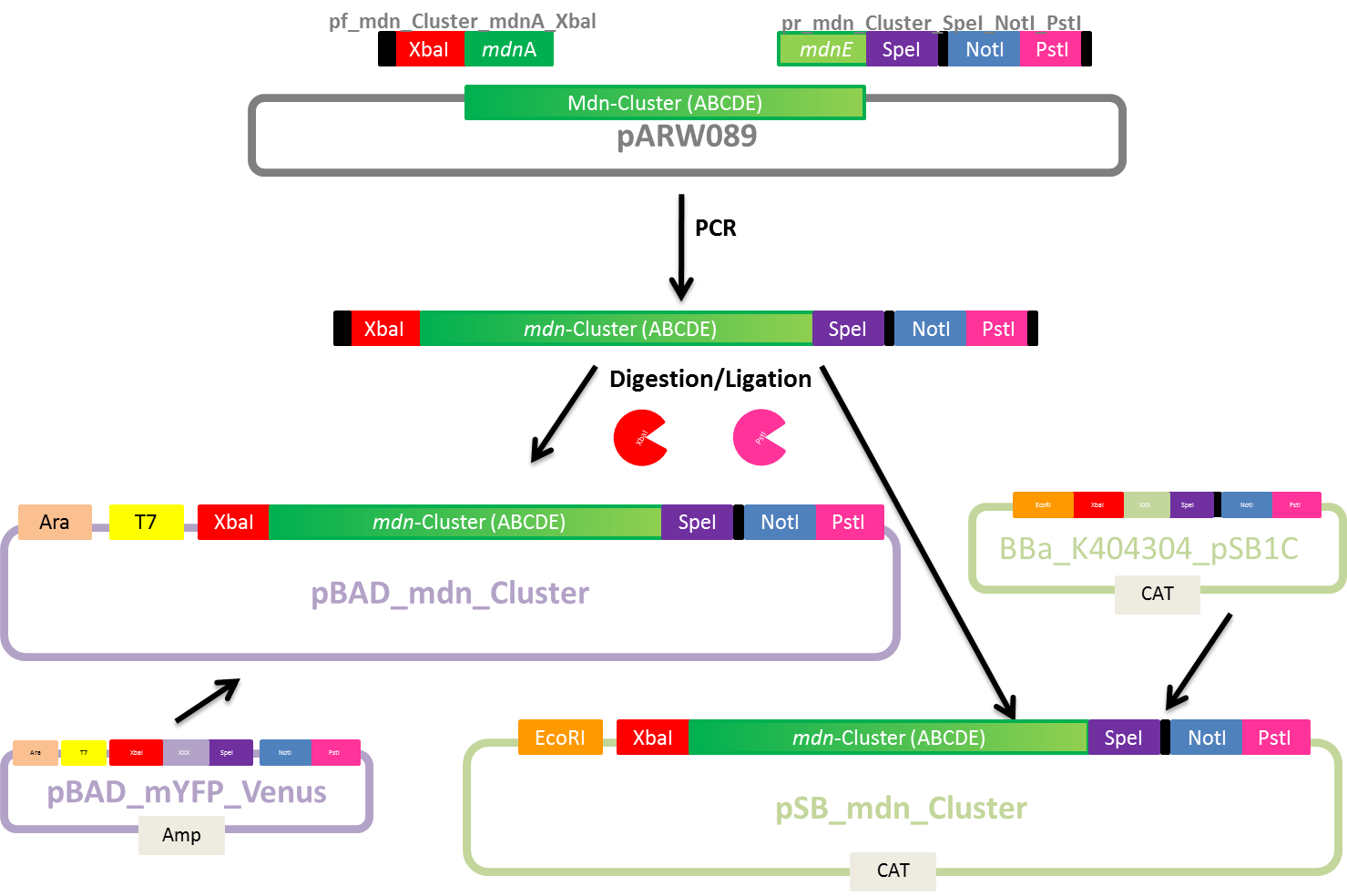
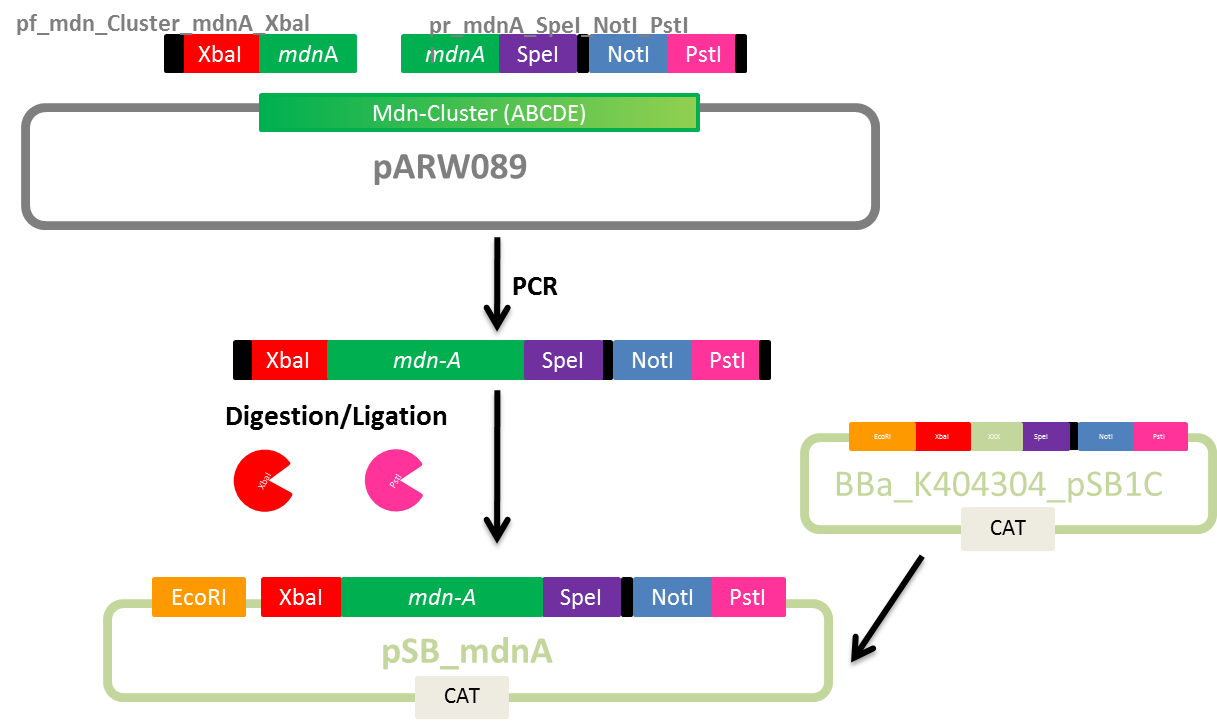
Design of Primers for BioBrick mdnA and mdn-Cluster
Investigators: Nicole, Nadine
Aim: Design primers for mdnA and mdn-Cluster(from pARW089) for BioBricks pBAD_mdn_Cluster, pSB_mdn_Cluster and pSB_mdnA
Time: 2011-07-19,14:00-17:00
Materials:
- Geneious Pro 5.1.7
Method:
- Primer design
- melting temperature based on 4+2 method (for mdnA-overlapping part)
- adding sequences for restriction sites (considering reading frame)
- Check self-complementarity and melting temperature using Oligo Calc
Plan:
- mdn-Cluster BioBricks
- mdnA BioBrick
Results:
Primer sequences
Further tasks:
- do PCR of mdnA
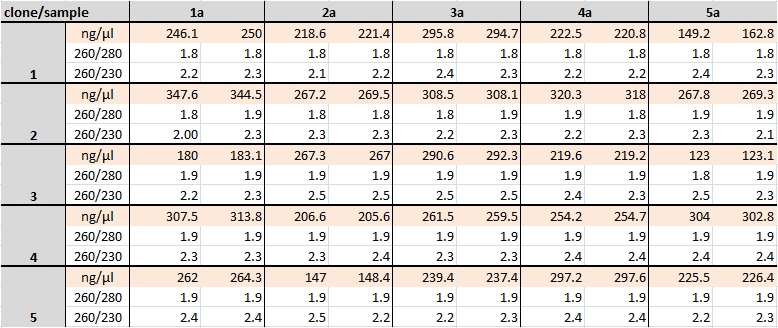
Miniprep of E. coli Xl blue containing mutated mndA in parW089
Investigators: Niels
Aim: 25x overnight cultures :
- miniprep
Time: 2011-07-19,12:00-16:00
Materials:
- E. coli Xl blue
Kit:
- Nucleic Acid and Protein Purification
- Support protocol for NucleoSpin Plasmid
Results:
Nanodrop:
Further task:
- sequencing
- digest
40th Labday 2011-07-20
Resistance test of the Dual-Transformation of XL1-blue (E. coli) with pTEV-SCS1 and pJC354_ssTorA_NheI_CS-TEV_XhoI_bla
Investigators: Stefan
Aim:
- overnight cultures for resistance test
Materials:
- Overnight cultures of
- XL1-blue transformed with pJC354_ssTorA_NheI_CS-TEV_XhoI_bla - growth with chloramphenicol in LB media
- XL1-blue transformed with pTEV-SCS1 and pJC354_ssTorA_NheI_CS-TEV_XhoI_bla - growth with kanamycin and chloramphenicol in LB media
Used method:
Restriction enzyme digestion pARW089 and pARW071
Investigators: Katharina, Nadine
Time: 2011-07-21,11:00-12:00
Aim: Restriction enzyme digestion of pARW089 and pARW071
Materials:
- Restriction enzymes: AatII, EheI (both from Elke)
- NEB Buffer 4
- Plasmids: pARW089, pARW071
Problem:
- AatII was empty (ordered new batch for tomorrow)
overnight culture of E. coli Xl blue m. ligation mdnA and geneIII into pARW089
Investigators: Sandrina
Aim: repeated test digestion of ligation mdnA and geneIII into pARW089 from 2011-07-12/13
Time: 16:30-18:00
Method/Materials:
- LB medium
Further tasks:
plasmid preparation and test digestion
Repeated PCR of mdnA and gene III for phage display (strategy 1+2)
Investigator: Sandrina
Time: 2011-07-26,9:00-11:00
Aim:
- amplification of mdnA with SfiI restriction sites (strategy 1)
- amplification of mdnA with NarI and AgeI and rfc 25 restriction sites (strategy 2)
- amplificate GeneIII with NgoMIV and AatII and rfc 25 restriction sites (strategy 2)
Primer:
- primer: pf_sfi-mdnA_2 and pr_sfi_mdnA_myc (mdnA, strategy 1)
- primer: pf_mdnA_iGEM_EheI and pr_mdnA_iGEM_AatII (mdnaA, strategy 2)
- primer: pf_geneIII_NgoMIV and pr_geneIII_iGEM_AatII (geneIII, strategy 2)
Reaction Components:
- 5 µl Vector pARW089
- 0,25 µl OneTaq Polymerase
- 1 µl dNTPs
- 1 µl per primer
- 5 µl 5x PCR Buffer
- 37,75 µl DNase free water
- purification of PCR fragments with QIAquick Gel Extraction Kit (250)
Further tasks:
- digestion of PCR products
digestion of PCR products and vectors (strategy 1+2)
Investigator: Sandrina
Time: 2011-07-26,11:00-13:00
Aim:
- digestion of mdnA and gene III for getting an mdnA-geneIII fusion gene with rfc25 restrition sites after ligation
- digestion of pARW089 for ligation of mdnA/geneIII fusion gene into it (strategy 2)
- digestion of pak100blaKDIR and and mdnA to enable ligation
Time: 2011-07-26,14:00-17:30
digestion enzymes:
- digestion of mdnA (strategy 2) with NarI and AgeI
- digestion of geneIII (strategy 2) with NgoMIV and AatII
- digestion of pARW089 (strategy 2) with NarI and AatII
- digestion of pak100blaKDIR (strategy 1) with SfiI
- digestion of mdnA (strategy 1) with SfiI
reaction components:
- 4 µl NEB 10x buffer
- 1 µl per restriction enzyme
- 30 µl PCR product / 5 µl vector
- 4 µl H²O
- BSA (only Sfi digestion)
reaction conditions:
- 1 h for PCR fragments
- 3 h for plasmids
- 50°C (SfiI digestion)
- 37°C NarI, AgeI, NgoMIV and AatII digestion
ligation of mdnA and geneIII into digested pARW089(strategy 2)
Investigators: Sandrina
Aim: get phage display vector pPDV089
Time: 2011-07-20,14:00-16:00
Materials:
- 6 µl digested vector pARW089 (NarI, AatII)
- 1 µl geneIII
- 1 µl mdnA
vector and insert should be added in ratio 1:3
- 2 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 9 µl water
- incubation over night at 16°C
Further Tasks: transormation of competent cells
ligation of mdnA into pak100blaKDIR (stategy 1)
Investigator: Sandrina
Aim: generate phage display vector pPDV100 (strategy 1)
Time: 2011-07-20,14:00-16:00
Method/Materials:
- 3 µl (ca 90 ng) Sfi-digested vector pak100
- 1 µl (ca 20 ng) Sfi-digested PCR fragment mdnA
- 2 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 13 µl water
- incubate over night at 16°C
Further Tasks: transformation of competent cells
41th Labday 2011-07-21
Test digestion of ligation for strategy 2
Investigators: Sandrina
Aim:repeated test digestion of ligation mdnA and geneIII into pARW089 from 2011-07-12/13
Time: 15:30-19:00
Materials/Methods:
Strategy 2:
- 0,5 µl XbaI
- 0,5 µl SpeI
- 2 µl 10x buffer 4 (NEB)
- 0,2 µl BSA
- 10 µl vector DNA
- 12,8 µl H2O
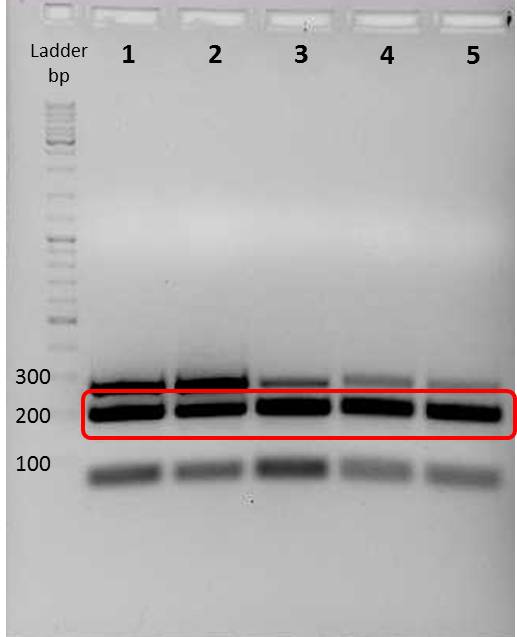
incubate for 1 h at 37°C
Results:
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | marker, DNA ladder mix, fermentas | 2 | |
| 1-20 | digested ligations | 6 | ca. 5000 bp, 4000 bp, 600 bp and 200 |
Further tasks:
sequencing of clone 6
Transformation of ligated pARW089 with mdnA-geneIII and ligated PAK100 bla KDIR with mdna
Investigators: Sandrina
Aim:Transformation of phage display ligations (strategy 1 and2) (2011-7-20) using heat shock
Time: 10:00-12:00
Method/Materials:
1) Take chemically competent E. coli XL1 blu cells from –80°C freezer.
2) Turn on heat block to 42°C.
3) use 60 µL of competent cells for each approach
4) Keep tubes on ice.
5) Add 2 µL DNA solution into the E. coli cell suspension, mix by flicking the tube. Incubate on ice for 15-30 min.
6) Put tubes into heat block at 42°C for 45 seconds.
7) Put tubes back on ice for 2 minutes to reduce damage to the E. coli cells.
8) Add 750 µL of LB with no antibiotic. Incubate tubes for 1 hour at 37°C and 750 rpm.
9) spin down the cell suspension for 3 min at 6000 rcf at 4 °C , discard most of the supernatant, resuspend the bacterial pellet by pipetting and use this for spreading.
10) Spread 100 ul of the resulting culture on LB agar plates with Tet/CM.
11) Grow overnight at 37 °C.
12) Pick colonies about 12-16 hours later.
Further tasks:
picking clones and plasmid preparation
42th Labday 2011-07-22
Restriction enzyme digestion pARW089 and pARW071 AND trouble-shouting
Investigators: Katharina, Nadine
Time: 2011-07-22,10:00-15:00
Aim: Restriction enzyme digestion of pARW089 and pARW071
Materials:
- Restriction enzymes: AatII, NarI
- NEB Buffer 4
- Plasmids: pARW089, pARW071
43th Labday 2011-07-23
Restriction enzyme digestion pARW089 (different incubation times), plasmid purification and ligation with EP-PCR mdnA
Investigators: Nadine
Time: 2011-07-22,14:30-20:30
Aim: Restriction enzyme digestion of pARW089 with different incubation times to test best digestion conditions, problems with the digestion of pARW089 in the last trials (-->gel excision and ligation if it worked out
Materials:
- Restriction enzymes: AatII, NarI, control: XmaI
- NEB Buffer 4
- PCR products of random mutated mdnA(1a-5a), purified from gel
- vector pARW089
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- GeneRuler 1kb DNA Ladder (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Digestion:
| components | volume of pARW089 /µl |
| DNA | 5 |
| NEB Buffer 4(10x) | 4 |
| Enzyme AatII | 0.8 |
| Enzyme NarI | 2 |
| H2O | 28.2 |
| Total volume | 40 |
2. Digestion: control
| components | volume of pARW089 /µl |
| DNA | 5 |
| NEB Buffer 4(10x) | 4 |
| Enzyme XmaI | 1 |
| BSA | 0.2 |
| H2O | 29.2 |
| Total volume | 40 |
- digestion at 37°C for 1 h
3. Production of agarose gel
- 0.8% gel for vector: mAgarose = 0.4 g in 50 ml 1x TAE adding 4 µl gel red
4. Loading samples
- 40 µl digest and 8 µl 6x loading dye
5. Run
- 110 V, time: >40 min
Results:
Gel
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | GeneRuler 1kb DNA Ladder | 2 | 10296, 116 |
| 1 | digested pARW089 1 hr | 48 | 10296, 116 |
| 2 | digested pARW089 2 hrs | 48 | 10296, 116 |
| 3 | digested pARW089 2.5 hrs | 48 | 10296, 116 |
| 4 | digested pARW089 XmaI 3 hrs | 48 | 3270, 7142 |
| 5 | digested pARW089 3 hrs | 48 | 10296, 116 |
| 6 | digested pARW089 undigested | 48 |
6. Gel excision
- lane 4 and 6, band: 10296, respectively
7. Ligation
Ligation: 3 µl vector (pARW089 digested with NarI and AatII, incubation 2.5 hrs and 3 hrs), 3 µl insert (mdnA digested with NarI and AatII from 2011-07-29), 1 µl T4 ligase, 1 µl T4 ligase NEB-buffer, overnight at 16°C
samples:
- pARW089 (dig. 2.5 hrs) + mdnA 1A
- pARW089 (dig. 2.5 hrs) + mdnA 2A
- pARW089 (dig. 2.5 hrs) + mdnA 3A
- pARW089 (dig. 2.5 hrs) + mdnA 4A
- pARW089 (dig. 2.5 hrs) + mdnA 5A
- pARW089 (dig. 3 hrs) + mdnA 1A
- pARW089 (dig. 3 hrs) + mdnA 2A
- pARW089 (dig. 3 hrs) + mdnA 3A
- pARW089 (dig. 3 hrs) + mdnA 4A
- pARW089 (dig. 3 hrs) + mdnA 5A
Further tasks:
transformation of E. coli cells(XL1-blue)
creation of pPDV089 in Genious
Investigators: Sandrina, Sabine
Aim: getting a model of pPDV089 for sequence alignment with purified pPDV089 plasmids from clones
Time: 09.00-11.00
Method/Materials:
software Genious
Further tasks:
perform alignment
sending pPDV089 from positive clone to GATC for sequencing
Investigators: Sabine
Aim: get sequence of generated phage display vector pPDV089 (strategy 2) to control the ligation of digested pARW089 with digested mdnA and gene III
Method/Materials:
10 µl of purified pPDV089 and 20 µl primer sf_mdna_1 were sent to GATC
Further tasks:
perform alignment
44th Labday 2011-07-24
Transform ligation from 42th Labday into E. coli XL blue 1 cells
Investigators: Stefan
Time: 2011-07-22,11:00-15:00
Aim: cells containing the desired vectors
Materials/Methods:
1)Take chemically competent E. coli XL1 blu cells from –80°C freezer.
2) Turn on heat block to 42°C.
3) use 60 µL of competent cells for each approach
4) Keep tubes on ice.
5) Add 2 µL DNA solution into the E. coli cell suspension, mix by flicking the tube. Incubate on ice for 15-30 min.
6) Put tubes into heat block at 42°C for 45 seconds.
7) Put tubes back on ice for 2 minutes to reduce damage to the E. coli cells.
8) Add 750 µL of LB with no antibiotic. Incubate tubes for 1 hour at 37°C and 750 rpm.
9) spin down the cell suspension for 3 min at 6000 rcf at 4 °C , discard most of the supernatant, resuspend the bacterial pellet by pipetting and use this for spreading.
10) Spread 100 ul of the resulting culture on LB agar plates with Tet/CM. 11) Grow overnight at 37 °C. 12) Pick colonies about 12-16 hours later.
45th Labday 2011-07-25
overnight culture of E. coli XL-1 blue containing mutated mndA in parW089
Investigators: Steffi
Aim: Picking 1 clone from each plate (Ligation pARW089 mdnA 2hrs 1A-5A, Ligation pARW089 mdnA 3hrs 1A-5A) and do an overnight culture
Time: 16:00-17:00
Method/Materials:
E. coli Strain: XL1 blue, Plasmid:mdnA in parW089
5 mL of LB media were supplied with 5 µL of Kan and cells were incubated at 37 °C overnight.
Further tasks:
plasmid preparation
46th Labday 2011-07-26
Miniprep of E. coli Xl blue containing mutated mndA in parW089
Investigators: Steffi
Aim: 10x overnight cultures :
- miniprep
Time: 2011-07-26,09:00-16:00
Materials:
- E. coli Xl blue
Kit:
- Nucleic Acid and Protein Purification
- Support protocol for NucleoSpin Plasmid
Further task:
- sequencing
Alignment of sequenced pPDV089 and model of pPDV089
Investigator: Sabine
Time: 2011-07-26,10:30-11:30
Aim: control of generated phage display vector pPDV089
Materials/Methods:
- file sent from GATC
- software Genious
Results:
- 2 mutations in geneIII
- the last 2 aminoacids of myc are absent
Further task:
- repeat PCR of all fragments using polymerase S (not OneTaq as before)
Repeated PCR of mdnA and gene III for phage display (strategy 1+2)
Investigator: Sabine
Time: 2011-07-26,11:30-13:30
Aim:
- amplification of mdnA with SfiI restriction sites (strategy 1)
- amplification of mdnA with NarI and AgeI restriction sites (strategy 2)
- amplificate GeneIII with NgoMIV and AatII restriction sites (strategy 2)
Primer:
- primer: pf_sfi-mdnA_2 and pr_sfi_mdnA_myc (mdnA, strategy 1)
- primer: pf_mdnA_iGEM_EheI and pr_mdnA_iGEM_AatII (mdnaA, strategy 2)
- primer: pf_geneIII_NgoMIV and pr_geneIII_iGEM_AatII (geneIII, strategy 2)
Reaction Components:
- 5 µl Vector pARW089
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl per primer
- 5 µl 10x PCR Buffer S
- 37,75 µl DNase free water
- purification of PCR fragments with QIAquick Gel Extraction Kit (250)
Further tasks:
- digestion
digestion of PCR products and vectors (strategy 1+2)
Investigator: Sabine
Aim:
- digestion of mdnA and gene III for getting an mdnA-geneIII fusion gene with rfc25 restrition sites after ligation
- digestion of pARW089 for ligation of mdnA/geneIII fusion gene into it (strategy 2)
- digestion of pak100blaKDIR and and mdnA to enable ligation
Time: 2011-07-26,14:00-17:30
digestion enzymes:
- digestion of mdnA (strategy 2) with NarI and AgeI
- digestion of geneIII (strategy 2) with NgoMIV and AatII
- digestion of pARW089 (strategy 2) with NarI and AatII
- digestion of pak100blaKDIR (strategy 1) with SfiI
- digestion of mdnA (strategy 1) with SfiI
reaction components:
- 4 µl NEB 10x buffer
- 1 µl per restriction enzyme
- 30 µl PCR product / 5 µl vector
- 4 µl H²O
- BSA (only Sfi digestion)
reaction conditions:
- 1 h for PCR fragments
- 3 h for plasmids
- 50°C (SfiI digestion)
- 37°C NarI, AgeI, NgoMIV and AatII digestion
Further Tasks:
- gel electrophoresis for purification of the digested fragments and vectors
gel electrophoresis of digested fragments and vectors
Investigator: Sabine
Aim: control PCR and digestion
Time: 2011-07-06,18:00-19:00
Results:
sizes of bands
- pak100blaKDIR (SfiI, strategy 1): ca 4,5 kp
- pARW089 (NarI and AatII, strategy 2): ca 10,5 kb
- mdnA (SfiI, strategy 1): ca 250 bp
- mdnA (NarI and AgeI, stategy 2): ca 200 bp
- geneIII (NgoMIV and AatII): no band
- purification with QIAquick Gel Extraction Kit (250)
Further Tasks:
- repeat PCR of geneIII
- ligation
Repeated PCR gene III for phage display (strategy 2)
Investigator: Sabine
Time: 2011-07-26,19:00-19:30
Aim:
- amplificate GeneIII with NgoMIV and AatII restriction sites (strategy 2)
Primer:
- primer: pf_geneIII_NgoMIV and pr_geneIII_iGEM_AatII (geneIII, strategy 2)
Reaction Components:
- 5 µl pak100blaKDIR
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl per primer
- 5 µl 10x PCR Buffer S
- 37,75 µl DNase free water
Further tasks:
- purification
- digestion
47th Labday 2011-07-27
digestion of PCR product geneIII (strategy 2)
Investigator: Sabine
Aim: digestion of gene III for getting an mdnA-geneIII fusion gene with rfc25 restrition sites after ligation
Time: 2011-07-26,09:30-10:30
reaction components:
- 4 µl NEB 10x buffer
- 1 µl per restriction enzyme (NgoMIV and AatII
- 30 µl PCR product
- 4 µl H²O
reaction conditions:
- 1 h, 37°C
Further Tasks:
- gel electrophoresis for purification of the digested geneIII
gel electrophoresis of digested geneIII
Investigator: Sabine
Aim: control PCR and digestion
Time: 2011-07-27,10:30-11:30
Method/Materials:
- 1% agarose gel
- GeneRuler DNA Ladder Mix (SM0331)
- 5x loading dye
- purification with QIAquick Gel Extraction Kit (250)
Results: size of band: ca 550 bp
Further Tasks: ligation
ligation of mdnA into pak100blaKDIR (stategy 1)
Investigator: Sabine
Aim: generate phage display vector pPDV100 (strategy 1)
Time: 2011-07-27,11:30-12:30
Method/Materials:
- 3 µl (ca 90 ng) Sfi-digested vector pak100
- 1 µl (ca 20 ng) Sfi-digested PCR fragment mdnA
- 2 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 13 µl water
- 1 h, room temperature
Further Tasks: transormation
Transformation of generated pPDV100 (strategy 1)
Investigators: Sabine
Aim:amplification of pPDV100
Time: 12:30-14:00
Method:
- addition of 10 µl ligation reaction to XL1-blue cells
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml chloramphenicol
- storage over night at 37°C
Further tasks:
control cell clones
48th Labday 2011-07-28
overnight culture of 10 picked clones of XL blue cells transformed with pPDV100
Investigators: Sabine
Aim: amplification and purification of generated phage display vector pPDV100 for test digestion and sequencing
Time: 10:00-10:30
Method/Materials:
5 ml LB medium per clone containining 100 µg/ml chloramphenicol
storage over night at 37°C and 800 rpm
Further tasks:
plasmid preparation, test digestion and sequencing
ligation of mdnA and geneIII (strategy 2)
Investigators: Sabine
Aim: get mdnA/geneIII fusion gene for cloning into pARW089
Time: 2011-07-28,11:00-12:30
Materials:
- 5 µl (ca 25 ng) digested geneIII (NgoMIV, AatII)
- 3 µl (ca 30 ng) digested mdnA (NarI, AgeI)
- 2 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 9 µl water
- 1 h, room temperature
Further Tasks: ligation of mdnA/geneIII into digested pARW089
ligation of mdnA/geneIII into digested pARW089(strategy 2)
Investigators: Sabine
Aim: get phage display vector pPDV089
Time: 2011-07-28,12:30-13:00
Materials:
- 6 µl (ca 70 ng) digested vector pARW089 (NarI, AatII)
- 1 µl (ca 5 ng) fusion gene mdnA/geneIII
- 2 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 10 µl water
- 1 h, room temperature
Further Tasks: transormation
Transformation of generated pPDV089 (strategy 2)
Investigators: Sabine
Aim: amplification of pPDV089
Time: 13:30-15:00
Method:
- addition of 10 µl ligation reaction to XL1-blue cells
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml tetracyclin and 100 µg/µl ampicillin
- storage over night at 37°C
Further tasks:
control cell clones
49th Labday 2011-07-29
Miniprep of E. coli overnight culture containing pPDV100
Investigators: Sabine
Aim: purification of pPDV100 for test digestion and sequencing
Time: 10:00-11:00
Method/Materials: see protocol 5.1 of the NucleoSpin Plasmid Kit
Further tasks: test digestion
gel electrophoresis of digested pPDV100
Investigator: Sabine
Aim: control ligated pPDV100
Time: 2011-07-29,11:30-12:30
Method/Materials:
1% agarose gel, GeneRuler DNA Ladder Mix (SM0331), 5x loading dye
Results: ligation of mdnA into pak100 has not worked
Further Tasks: control all working steps, repeat cloning
creation of pPDV100 in Genious
Investigators: Sandrina, Sabine
Aim: getting a model of pPDV100 for sequence alignment with purified pPDV089 plasmids from clones
Time: 14.00-16.00
Method/Materials: software Genious
Results: subsequent insertion of myc into reverse PCR primer (mdnA) caused frameshift, new primer ordered
 "
"