Team:Duke/Project
From 2011.igem.org
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling and Results | Notebook | Safety | Attributions |
|---|
Contents |
Overall project
Additions to the synthetic biologists' toolkit are expected to be interoperable and modular in order to facilitate standardization of these tools. The Genetic Toggle Switch originally presented by Gardner et al (2000) was a major breakthrough in synthetic biology and utilized constitutive promoters. Here, we use Zinc Fingers as transcriptional repressors in a newly designed interfacing network, the controller, for a modified version of the original toggle switch. The controller interface is made with emphasis on interoperability and applications in mind. Specifically, we use zinc finger repressors and degradation tags to reduce cross-talk and make the network more robust. Nine ZF transcription factors are computationally characterized and submitted to the registry. The interfacing controller and toggle switch are stochastically modeled to predict gene expression levels and are subjected to experimental testing.
Project Details
Introduction to Genetic Toggle Switches
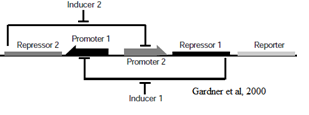
The Genetic Toggle Switch was among the first artificially constructed Gene Regulatory Networks. The original Genetic Toggle Switch consisted of the ptrc-2 (lacI repressible) promoter paired with the temperature sensitive lambda repressor and the PLs1con (lambda repressible) promoter paired with the lacI repressor. Subsequently, induction of the ptrc-2 promoter would repress the PLs1con promoter through expression of the lambda repressor, while induction of the PLs1con promoter would repress the ptrc-2 promoter through expression of the lacI repressor. Switching of the toggle switch from one promoter to another was accomplished by addition of either Isopropyl β-D-1-thiogalactopyranoside (IPTG), which causes the lacI to unbind allowing the ptrc-2 to switch on, or a thermal pulse, which causes the lambda repressor to unbind and allow the PLs1con promoter to switch on. In order to characterize the toggle switch, a GFPmut3 structural gene was placed downstream of the Ptrc-2 promoter so that induction of the Ptrc-2 promoter would result in high expression of GFPmut3 while induction of PLs1con would result in low expression of GFPmut3.
Characteristics of Genetic Toggle Switches
In general, Genetic Toggle Switches have several characteristics which make them unique from other Gene Regulatory networks. For example, Genetic Toggle Switches must be capable of bi-stable behavior, meaning that they should only exist in one of two stable states instead of in a variety of intermediate states. Bi-stability also suggests that a certain threshold must be passed before the toggle switch can switch from one state to another. In addition, toggle switches should have low basal transcriptional noise to prevent random switching without outside induction. Finally, a reporter or marker structural gene, such as a fluorescent protein, should be present in order to characterize the effectiveness of the toggle switch.
Design of Zinc Finger Transcription Factors
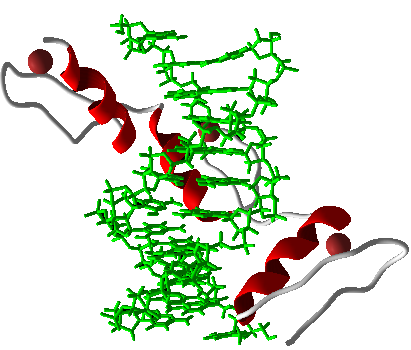
Zinc Fingers (ZFs) are proteins serving as DNA binding domains with high binding specificity and affinity. ZFs also have the ability to form 3-finger arrays . Modular assembly of ZFs regards the binding of individual ZF modules to produce multi-finger arrays (ZFAs) and requires the creation of a randomized library of ZFs with arbitrary binding sites (Joung et al, 2000). After analysis of modified modular assembly , it was determined some ZFAs, made with Context dependent Assembly (CoDA), had greater specificity and affinity with certain module pairings present in the array (Ramirez et al, 2008). We assembled 9 zinc finger transcription factors using the CoDA method and the computational program, ZiFit. These nine zinc fingers each bound to a nine base pair sequence that occurred no more than 2 times in the natural E. coli genome. In order to confirm that each 9-bp binding site occurred no more than twice we used BLASTn to search the E. coli genome for any potential binding sites. A low level of binding sites is desirable because it guarantees minimal interference between synthetic and natural parts.
Zinc Finger Transcription Factors
The use of Zinc Finger Transcription factors (TFs) in eukaryotic systems has been developed (Beerli et al, 2002). However, the use of characterized zinc finger TFs with specified DNA Binding Sites (DBSs) in prokaryotes in was recently shown to have the potential to revolutionize the exploration of GRNs and synthetic biology as a whole (Holtz, 2011 unpublished work). The power of ZFs as a DNA binding domain was tagged with various repression techniques such as and steric hindrance of RNA polymerase (RNAP) yielded effective transcriptional repressors of synthetic promoters (Holtz, 2011 unpublished work). This advance in the construction of synthetic repressor-promoter pairings has set the stage for major breakthroughs and increased efficiency in the field.
Network Design
A variety of Network designs using zinc finger transcription factors were considered before we settled on the Genetic Toggle Switch. Some of the rejected designs included novel uses of AND/OR gates to create a "bacterial encryption security" system. Network design began with simple white board drawings before moving on to more sophisticated drawing and modeling software.
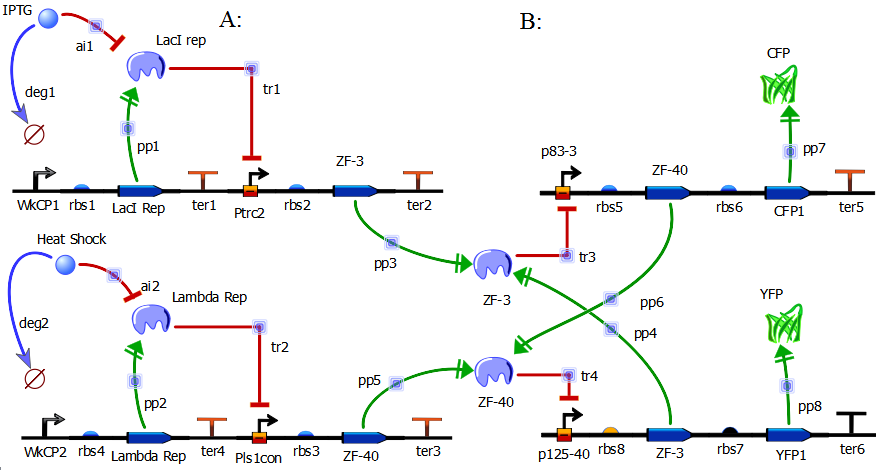
In redesigning the genetic toggle switch we sought to retain the following important characteristics of the original toggle switch.
-Bi-stability
-Low Basal Transcriptional Noise
-Toggling via Chemical Inducers
-Usage of Constitutive Repressible Promoters
-Usage of fluorescent proteins to indicate stable states
In order to redesign the genetic toggle switch we used a variety of programs including ApE (A plasmid Editor) to string together the sequences for our plasmids and TinkerCell to create a diagram of our network as well as to model the network dynamics using stochastic differential equations.
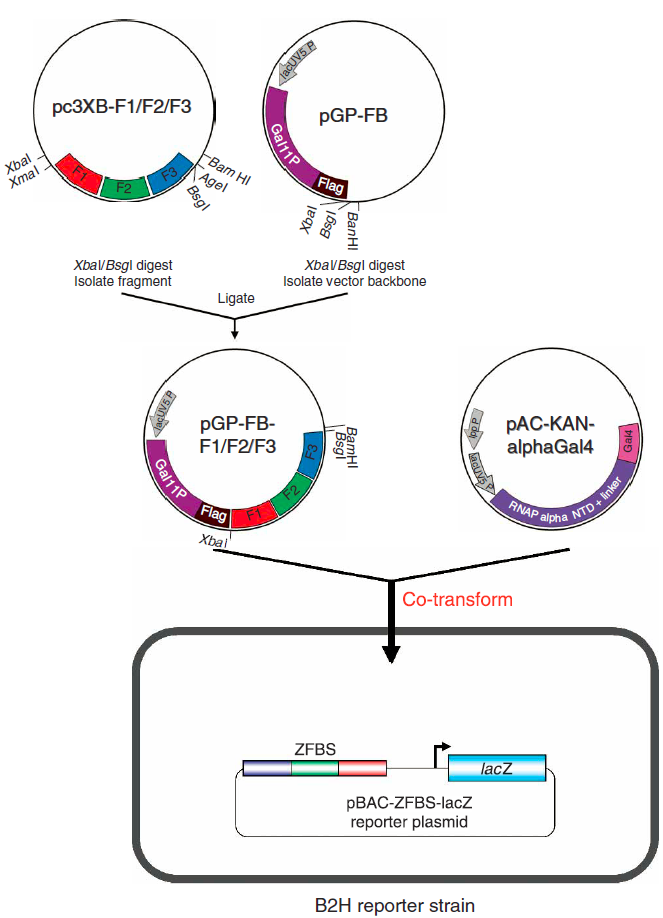
Bacterial 2-Hybrid Assay
Zinc finger transcription factors were characterized in vivo via the bacterial two hybrid (B2H) assay. In this assay, selected zinc finger arrays, which were synthesized de novo, were first transformed into transcriptional activators through the addition of an activation domain. These transcriptional activators then promote expression of the lacZ reporter gene, which is used to determine binding affinity via fold-activation measurements. Furthermore, this assay presented a systematic method to test zinc fingers as transcription factors within the same expression system we planned to implement our network design in. We primarily followed the procedure as described by Wright et al (2006). However, we adjusted this protocol to take into account the restriction sites flanking the sequences generated by ZiFiT. The restriction enzyme used to make the second cut in the expression vector and zinc fingers, in steps 25-49 of the assay, was changed from BsgI to BamHI.
CPEC Cloning
Materials
Phusion™ High-Fidelity PCR Kit (FINNZYMES, Cat. No. F-553)
Thermocycler
Preparation
5x Phusion HF Buffer 4 ul
10 mM dNTPs 0.4 ul
Vector 50 ng/1kb
Insert x ng*
Phusion DNA Polymerase 0.2 ul
H2O to 20 ul
- The amount of insert is determined so that the molar ratio for vector and insert is 1 to 2.
Procedures 98°C 30sec
10X
98°C 10 sec
Annealing** 30 sec
72°C x sec***
72°C 5min 4°C hold
- Anneal at Tm + 3°C. The Tm should be calculated with the nearest-neighbor method.
- The extension time is usually calculated according to the shortest piece with 15 sec /kb if the cloning is not complicated. For example, if there is only one insert and is shorter than the vector, say, 600 bp, then I will use 15 sec for extension. Refer to the published paper for detailed information.
- Anneal at Tm + 3°C. The Tm should be calculated with the nearest-neighbor method.
DNA Purification Materials : E.Z.N.A Gel Purification Kit (Omega Bio-Tek, Cat No. D2500-02 ) Water bath equilibrated to 55-65C Microcentrifuge capable of at least 10,000 x g Nuclease-free 1.5 ml centrifuge bottles Absolute (95%-100%) ethanol Protective eye-wear Isopropanol (for fragments < 500 bp only) Protocol: 1. Perform agarose gel electrophoresis to fractionate DNA fragments. Any type or grade of agarose may be used. It is strongly recommended, however, that fresh TAE buffer or TBE buffer be used as running buffer. Do not re-use running buffer as its pH will increase and reduce yields. 2. When adequate separation of bands has occurred, carefully excise the DNA fragment of interest using a wide, clean scalpel. 3. Determine the approximate volume of the gel slice by weighing it in a clean 1.5 ml microfuge tube. Assuming a density of 1 g/ml of gel, the volume of gel is derived as follows: A gel slice of mass 0.3 g will have a volume of 0.3 ml. Add equal volume of Binding Buffer (XP2). Incubate the mixture at 55C-60C for 7 min or until the gel has completely melted. Mix by shaking or inverting the tube every 2-3 minutes. Centrifuge the tube briefly to collect all the liquid to the bottom of the tube. Note: For DNA fragment less than 500bp, add 1 sample volume of isopropanol after the addition of Binding Buffer (XP2). 1. Apply up to 700 ul of the DNA/agarose solution to a HiBind® DNA spin column assembled in a clean 2 ml collection tube (provided) and centrifuge in a microcentrifuge at 8,000-10,000 x g for 1 min at room temperature. Discard the liquid. Re-use the collection tube in Steps 5-8. For volumes greater than 700 ul, load the column and centrifuge successively, 700 ul at a time. Each HiBind® spin-column has a total capacity of 25-30 ug DNA. 2. Discard liquid and add 300ul Binding Buffer. Centrifuge at 10,000 x g for 1 minutes. 3. Add 700 ul of SPW Buffer diluted with absolute ethanol into the column and wait 2-3 minutes. Centrifuge at 10,000 x g for 1 min at room temperature to wash the sample. 4. Discard liquid and repeat step 6 with another 700 ul SPW Buffer. 5. Discard liquid and, re-using the collection tube, centrifuge the empty column for 1 min at maxi speed (>13,000 x g) to dry the column matrix. This drying step is critical for good DNA yields. 6. Place column into a clean 1.5 ml microcentrifuge tube (not provided). Add 30-50 ul depending on desired concentration of final product) Elution Buffer (or sterile deionized water) directly to the center of the column matrix, then incubate for 1 minute. Centrifuge 1 min at maxi speed (>13,000 x g) to elute DNA. This represents approximately 70% of bound DNA. PCR Materials Phusion™ High-Fidelity PCR Kit (FINNZYMES, Cat. No. F-553) Thermocycler Preperation 5x Phusion HF Buffer 10 ul 10 mM dNTPs 1 ul DNA template 1 pg – 10 ng Forward primer (10 uM) 2.5 ul Reverse primer (10 uM) 2.5 ul Phusion DNA Polymerase 0.5 ul
-----
H2O to 50 ul
Procedure: 98°C 30sec
30X 98°C 10 sec Annealing* 30 sec 72°C 15 sec per 1 kb
72°C 5min 4°C hold
Transformation:
Materials:
TOP10 Chemical Competent Cells (Invitrogen, Cat No. C4040-03)
SOC Medium (Sigma, Cat. No. S1797)
LB Agar (Sigma, Cat. No. L3027)
Petri Dishes (VWR, Cat. No. SC25373-187)
Cell Spreader (VWR, Cat. No. 89042-018)
37°C incubator
37°C shaker
water bath
Protocol:
1. Thaw 1 tube of competent cells on ice;
2. Add 3 ul of cloning product or 1-50 ng of plasmid into competent cells while stirring gently;
3. Keep the tube covered by ice for 30min;
4. Heat-shock the competent cells in water bath for 45 sec at 42°C;
5. Put the tube on ice for 2 minutes;
6. Add 450 ul of SOC medium and then put it in a 37°C shaker for 1 hour;
7. Dilute and spread an appropriate amount on an LB agar plate with the appropriate antibiotics;
8. Place the plate up-side-down in 37°C incubator for 16-18 hours (overnight).
 "
"