Team:Washington/Magnetosomes/GibsonVectors
From 2011.igem.org
(→About Gibson Cloning/Assembly) |
(→What happened last year?) |
||
| Line 13: | Line 13: | ||
=== What happened last year?=== | === What happened last year?=== | ||
The Gibson Cloning method is definitely not a new method to be introduced to the iGEM community. | The Gibson Cloning method is definitely not a new method to be introduced to the iGEM community. | ||
| - | In 2010, the Cambridge iGEM team created the [http://www.cambridgeigem.org/RFC57.pdf RFC 57] document which outlines a protocol for Gibson Assembly using | + | In 2010, the Cambridge iGEM team created the [http://www.cambridgeigem.org/RFC57.pdf BBF RFC 57] document which outlines a protocol for Gibson Assembly using standard BioBricks (BBF RFC 10) that would allow many fragment inserts during a single cloning step. However, while creating the Magnetosome Toolkit, we found that this BioBrick standard was incapable of producing high yields of desired Gibson products even for two-fragment assemblies. |
<br> | <br> | ||
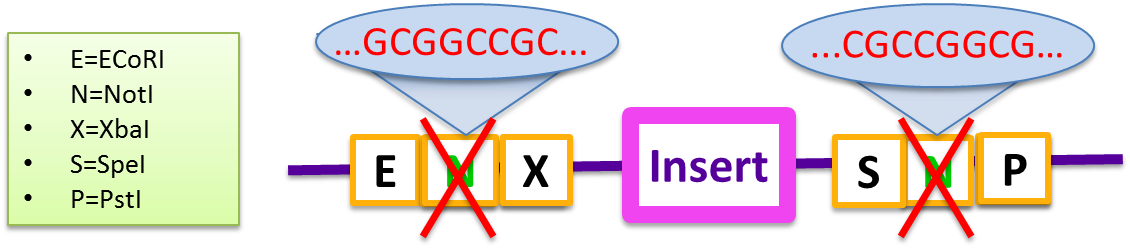
| - | The primary problem with | + | The primary problem with the standard pSB vector is the self-complementarity of the two NotI sequences embedded in both the BioBrick prefix and suffix. These sequences prevent gene inserts from being incorporated efficiently, and do not produce a high yield of the Gibson product even in two-fragment assemblies. Generally, the backbone self-anneals and the recircularized plasmid has a combined prefix and suffix region that reads EcoRI-NotI-PstI. <br/> |
[[File:Igem2011 biobrick NotI.png|600px|center]] | [[File:Igem2011 biobrick NotI.png|600px|center]] | ||
| - | + | To overcome this problem, the 2011 UW iGEM team developed new prefix and suffix regions that are based on BglBrick standards and designed to eliminate self-complimentarity of the plasmid. These vectors drastically increase the Gibson assembly efficiency of large-scale gene assemblies and are also compatible with iGEM standard BioBrick parts. For experimental details on the compared efficiencies of pSB1A3 and pGA1A3, see [https://2011.igem.org/Team:Washington/Magnetosomes/GibsonResults our assay results].<br/> | |
[[File:Igem2011_gibsonbrick.png|600px|center]] | [[File:Igem2011_gibsonbrick.png|600px|center]] | ||
Revision as of 22:16, 28 September 2011
About Gibson Cloning/Assembly
Gibson Cloning/Assembly is a novel synthetic biology tool that allows multiple gene-inserts during a single isothermal reaction that is used for assembling overlapping DNA fragments. This method is gaining popularity as it tends be more efficient, saving a great amount of time during the cloning process. Overall, Gibson cloning allows teams to built large gene constructs with ease.
What happened last year?
The Gibson Cloning method is definitely not a new method to be introduced to the iGEM community.
In 2010, the Cambridge iGEM team created the [http://www.cambridgeigem.org/RFC57.pdf BBF RFC 57] document which outlines a protocol for Gibson Assembly using standard BioBricks (BBF RFC 10) that would allow many fragment inserts during a single cloning step. However, while creating the Magnetosome Toolkit, we found that this BioBrick standard was incapable of producing high yields of desired Gibson products even for two-fragment assemblies.
The primary problem with the standard pSB vector is the self-complementarity of the two NotI sequences embedded in both the BioBrick prefix and suffix. These sequences prevent gene inserts from being incorporated efficiently, and do not produce a high yield of the Gibson product even in two-fragment assemblies. Generally, the backbone self-anneals and the recircularized plasmid has a combined prefix and suffix region that reads EcoRI-NotI-PstI.
To overcome this problem, the 2011 UW iGEM team developed new prefix and suffix regions that are based on BglBrick standards and designed to eliminate self-complimentarity of the plasmid. These vectors drastically increase the Gibson assembly efficiency of large-scale gene assemblies and are also compatible with iGEM standard BioBrick parts. For experimental details on the compared efficiencies of pSB1A3 and pGA1A3, see our assay results.
What about this year?
Seeing that this is a very efficient method to do cloning, we continued to make improvements to the methods and created a Gibson Assembly Toolkit!
Creation of 5 plasmid vectors
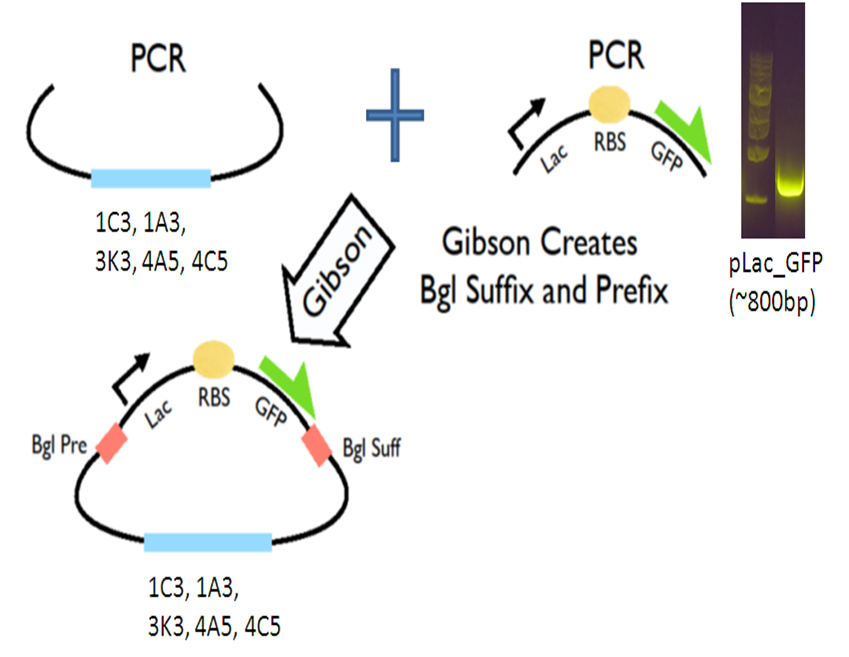
Our new vectors for Gibson assembly follow the naming convention of pGA. To make our pGA vectors, we first amplified the backbones and the pLac GFP insert respectively. Then we performed a Gibson reaction to combine them together to make the pGA vectors.
All togeher, we created 5 pGA vectors and submitted them to the registry:
- 2 High copy extraction/cloning vectors
- pGA1A3, pGA1C3
- 1 medium copy expression vector
- pGA3K3
- 2 low copy expression vectors
- pGA4A5, pGA4C5
As listed above, each of our vectors have varying copy numbers, antibiotic resistances, and purposes within the magnetosome gene assembly. However, they all appear to be very efficient will multiple gene inserts.
Wonder about the efficiency of the pGA vectors? Check out our evaluation report!
References
1.
 "
"