Design
The [http://www.nature.com/nature/journal/v463/n7279/abs/nature08753.html paper] our system is based on used microfluidic devices to measure oscillations. Such microfluidic devices however are very expensive and our iGEM team budget was limited, so we decided to design a customary fluidic device, which was produced in the Wageningen University workshop. To keep as many options as possible open, the design implemented the idea of potentially accommodating two bacteria growing platforms, a micro-sieve and a micro-dish. Gaining more in-depth understanding of our system, we decided to focus on the micro-dish as platform to test the oscillatory behaviour of our construct. Figure 1 and 2 below show the flow chamber as original scetch designed by our team and the 3D rendered depiction of the final device.

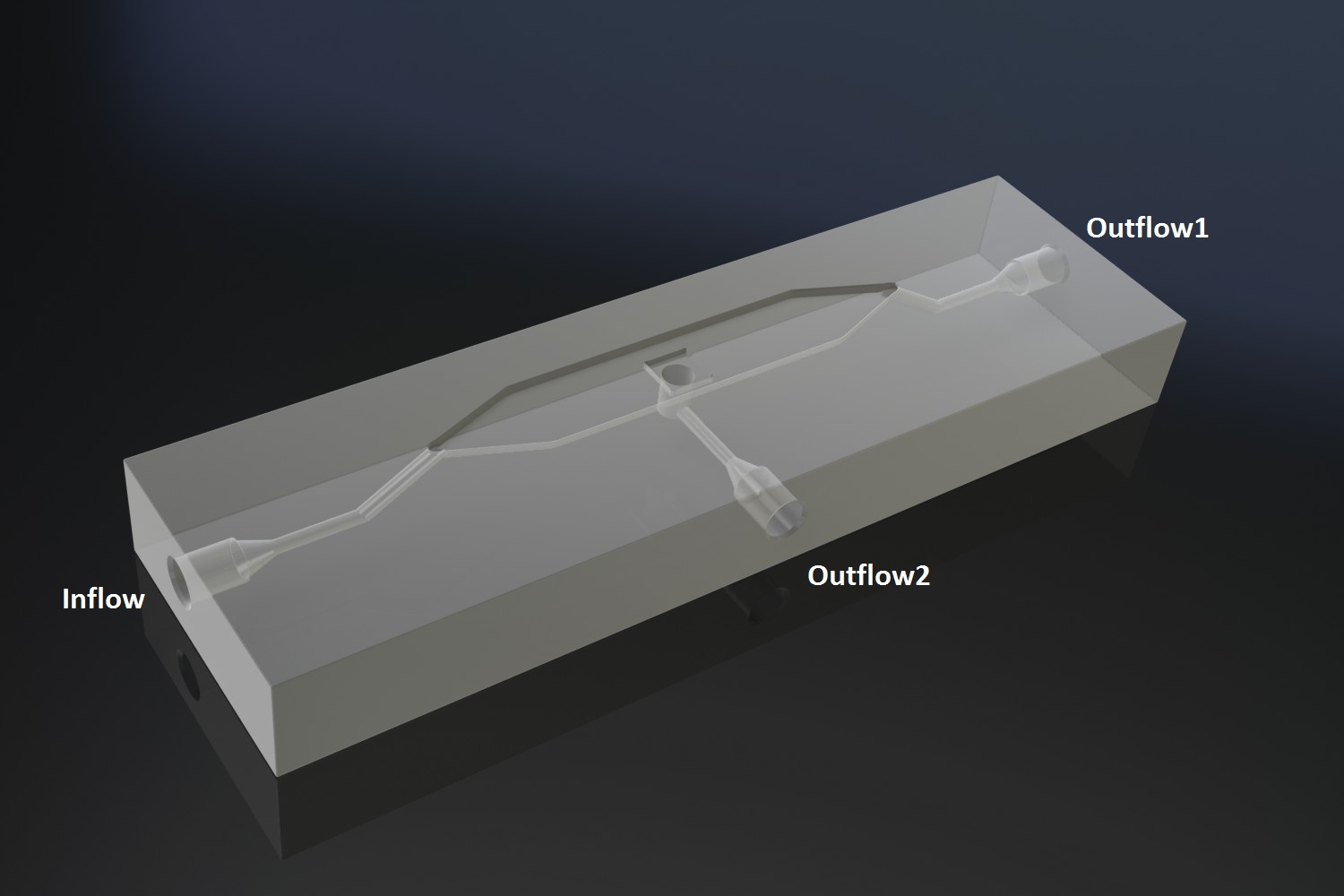
Fig.1: Design of the fluidic device: original scetch and 3D render
The micro-dish contains porous wells, in which the cells can be physically constrained. It is placed in the socket of the dish, which allows a fluid to be flown over the wells. This fluid can be used to supply the cells with nutrients and wash away excess AHL and cells if necessary. Figure 2 shows a schematic of the device. The inflow and outflow1 can be used to flush the device with fresh medium. The use of outflow2 depends on the bacteria growing platform. In the case of the micro-sieve it can be used to create an under pressure, e.g. with a syringe, to trap the bacteria on the sieve. When using a micro-dish, it can optionally be used to bottom-feed the bacteria in the wells and/or supply additional substances to initalize the oscillatory behaviour of the system, e.g. IPTG in the case of the pRight/lacI hybrid promoter for the streamligned oscillator.
Fig.2: Schematic of the micro-fluidic device depicting the position of in- and outflow
 Left: Production of the device in the Wageningen University workshop
Left: Production of the device in the Wageningen University workshop
back to top
 "
"






