Team:Brown-Stanford/REGObricks/Biocementation
From 2011.igem.org
Biocementation
Space and Biocementation
Current applications of urease activity include: concrete repair 1, limestone protection2, biogrouting 3 and increasingly, biocementation 4 5. Just in 2010, there was a multitude of publications from the Netherlands on the sciences of geoengineering, focusing explicitly on ureolytic bacteria as a suitable tool for repairing cracks in cement buildings and historical statues. Of course, our project seeks to expand the use of biocementation beyond just earthly affairs. As professor from UC. San Diego notes, “Water, and cement make good shielding but they are very heavy” (Prof. Craig Patten). We hope to alleviate this burden of extra weight for supplies by using S. pasteurii’s biocementation to create structures on-site.
What is Biocementation?
“Biocementation” is the growing field of employing biological agents as catalysts for increasing the stiffness and strength of soil [1]. The particle branch of biocementation that we investigated in our project this summer was “Microbial Induced Carbonate Precipitation (MICP)”, a process by which microorganisms metabolize various nitrogen products to increase ambient pH. The salient organism in our case is Sporosarcina pasteurii (previously Bacillus pasteurii), which utilizes the hydrolysis of urea to generate carbon dioxide and ammonia, which ultimately convert to carbonate ions in a basic environment. Carbonate fuses with available metals in solution to precipitate in crystal formation, and fuse surrounding particles together.
The Chemical Reaction
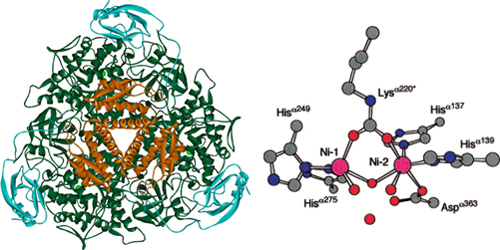
Urease (urea amidohydrolase; EC 3.5.1.5) is a six-subunit protein assembled from three alpha and three beta subunits.
Contained in the center of the urease enzyme is dual nickel metallocore, crucial to its activity 6. During each cycle of activity, urease breaks down one urea molecule into one molecule of ammonia and one molecule of carbamate, which is unstable and decomposes to another molecule of ammonia and bicarbonate. Each of the ammonia molecules have a pKa of 9.31, and take up protons from the cytoplasm. They are then shuttled out of the cell to create an proton gradient for ATP-generation. Outside the cell, ammonium contributes to an increase of pH in the surrounding environment. The cell itself also serves as a nucleation site, as positively charged calcium ions in solution are attracted by its negative membrane. The gradual evolution of carbonate from the basic environment begins the process of calcium precipitation, and the rest, they say, can be seen in stone!
The genetic sequence of the urease has been ascertained for several strains of bacteria, including Helicobacter pylori, and some partial mappings have been done for individual subcomponents of S. pasteurii 7,8. There have also been occasional mentions of strains of recombinant bacteria containing urease function 9 though unfortunately, none currently in a biobricked form. While the study of urease function in H. pylori has matured under medicinal interest over the years, the genetic expression of urease is still a field of ongoing research. Ambitiously, we decided that one of the project goals for REGObricks would be to curate an modular, enzymatically functional urease to the iGEM registry of Standardized Parts.
Check out how we did here.
More about Urease
Iconic in the history of protein science, urease was the first enzyme to be purified by James Sumner from a jack bean plant in 1926 [2]. Since then, urease has been well-known in the medical field as a convenient way to test for the presence of urinary tract pathogenetic bacteria such as Helicobacter pylori, one of the chief culprits in the formation of duodenal ulcers and stomach cancer. The strength and enzymatic activity of the enzyme is distributed across a wide range of species, reflecting the different functions it serves. A survey of some representative species of bacteria is given in Figure 1. Note S. pasteurii'’s sustentative range of urease activity.
Continue to our characterization of S. pasteurii!
References
1 Jonkers et al. Application of bacteria as self-healing agent for the development
of sustainable concrete. Ecological Engineering 36, (2010) pp.230–235.]]
2 Muynck, W. D. et al. Influence of urea and calcium dosage on the effectiveness of bacterially induced
carbonate precipitation on limestone. Ecological Engineering 36 (2010) 99–111]]
3 Van Paassen, Leon. Biogrout. Biogrout: Microbially Induced Carbonate Precipitation. BioGeoEngineering Conference. Web. 27 June 2011.]]
4 DeJong, J. et al. Bio-mediated soil improvement. Ecological Engineering 36, (2010) pp.197–210.]]
5 Harkes, Marien P. et al. Fixation and distribution of bacterial activity in sand to induce
carbonate precipitation for ground reinforcement Ecological Engineering 36 (2010) 112–117]]
6 Mobley HL, Island MD, Hausinger RP. (1995) Molecular biology of microbial ureases. Microbiol Rev 59.3 451–480. http://www.ncbi.nlm.nih.gov/pubmed/7565414 PubMed
7 Weeks DL, Eskandari S, Scott DR, Sachs G. (2000) A H+-Gated Urea Channel: The Link Between Helicobacter pylori Urease and Gastric Colonization. Science, 287 482-485.]]
8 Cruz-Ramos, H., P. Glaser, L. V. Wray, Jr., and S. H. Fisher. 1997. The Bacillus subtilis ureABC operon. J. Bacteriol. 179:3371-3373. [PMC free article] [PubMed].]]
9 Cussac V, Ferrero RL, Labigne A. (1992) Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol., 174 2466-2473.]]
 "
"