Team:HokkaidoU Japan/Project/Backbone
From 2011.igem.org
m |
|||
| (7 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div id="hokkaidou-right-content"> | <div id="hokkaidou-right-content"> | ||
| - | ==Bsa I | + | ==Ready-to-inject backbone and Bsa I cloning site== |
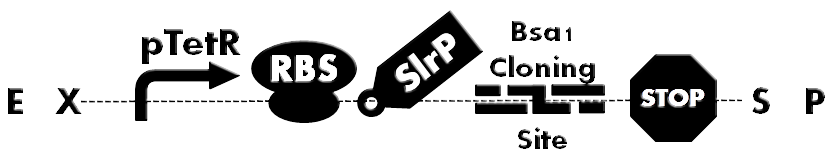
| - | [[File: | + | [[File:HokkaidoU_BsaI_Backbone.png|thumb|500px|Figure1. A backbone under constitutive promoter(pTetr). Has SlrP as a injection signal, GSK tag, Bsa I Cloning Site. Desired protein can be inserted into the cloning site.]] |
| - | Bsa I Cloning site has unique characteristics that | + | Bsa I Cloning site has unique characteristics that enabled us to clone BioBrick in to two flanking Bsa I restriction sites arranged in opposite directions and still retain whole constructs BioBrick properties. Cloning site was added downstream of SlrP region for construction of our backbones for T3SS characterisation. Bsa I cloning site is invaluable part when you need to repeatedly replace particular domain part at the middle of the construct. |
| - | Bsa I restriction enzyme | + | Bsa I restriction enzyme is classified as Type IIs restriction endonuclease. The unique property of this class is that recognition site is apart of restriction site . Unlike EcoR I or Pst I, Bsa I recognises GGTCTC sequence, but cuts the sequence located 7 bases downstream from first base recognised by Bsa I. Which results in a 5 prime 4 base overhang structure (Fig. 2). This is a key property that enables insertion of BioBrick in the middle of construct possible. |
<pre> | <pre> | ||
| Line 17: | Line 17: | ||
</pre> | </pre> | ||
| - | Of course there are other restriction endonucleases that exhibit same properties | + | Of course there are other restriction endonucleases that exhibit same properties. Using other enzymes of this class it is possible to add additional cloning sites in the same construct. |
| - | + | Designing the Bsa I a cloning site that the digestion would result in Not I like overhang and Spe I like overhang flanking the cloning site.(Fig. 3). | |
<pre> | <pre> | ||
| Line 34: | Line 34: | ||
</pre> | </pre> | ||
| - | Please | + | Please be careful Bsa 1 is not official BioBrick enzyme so you must check your plasmid backbone sequence and remove it if there is one . However you don have to worry about insert BioBricks, because they only need to be digested by official BioBrick enzymes. |
| + | |||
| + | For domain fusion, removal of existing stop codons of prefix and/or suffix is essential. This can be easily achieved by designing primers which delete stop codons by adding single point mutation. These primer sets can be used as universal primers which aneal to all BioBricks. | ||
| + | |||
| + | ==RFC submission== | ||
| + | We have submitted this method as [[Media:HokkaidoU_BBF_RFC_87.pdf|BBF RFC 87]]. | ||
| + | For more details about RFC submission, please see also [[Team:HokkaidoU_Japan/Project/RFC87|here]]. | ||
| - | |||
</div> | </div> | ||
{{Team:HokkaidoU_Japan/footer}} | {{Team:HokkaidoU_Japan/footer}} | ||
Latest revision as of 12:38, 15 December 2011
HokkaidoU Japan
iGEM 2011 Team of Hokkaido University
Contents |
- Abstract
- What`s T3SSDetailed information about T3SS and summary of our achievements on iGEM 2010
- Injection assay using onion cellsExperiments using plant cells are easier to perform than with mammalian ones
- Ready-to-inject backbone and Bsa I cloning siteReady-to-inject backbone and Bsa I cloning site enables easy fusion of T3S signal and protein
- GSK tag systemA neat injection assay using GSK tag, which can specifically detect successfully injected proteins
- Bsa I cloning site, RFC submissionDetailed documentation of costructing a BioBrick cloning site a BioBrick!
Ready-to-inject backbone and Bsa I cloning site
Bsa I Cloning site has unique characteristics that enabled us to clone BioBrick in to two flanking Bsa I restriction sites arranged in opposite directions and still retain whole constructs BioBrick properties. Cloning site was added downstream of SlrP region for construction of our backbones for T3SS characterisation. Bsa I cloning site is invaluable part when you need to repeatedly replace particular domain part at the middle of the construct.
Bsa I restriction enzyme is classified as Type IIs restriction endonuclease. The unique property of this class is that recognition site is apart of restriction site . Unlike EcoR I or Pst I, Bsa I recognises GGTCTC sequence, but cuts the sequence located 7 bases downstream from first base recognised by Bsa I. Which results in a 5 prime 4 base overhang structure (Fig. 2). This is a key property that enables insertion of BioBrick in the middle of construct possible.
Fig. 2 5'...GGTCTCN^.......3' 3'...CCAGAGNNNNN^...5'
Of course there are other restriction endonucleases that exhibit same properties. Using other enzymes of this class it is possible to add additional cloning sites in the same construct.
Designing the Bsa I a cloning site that the digestion would result in Not I like overhang and Spe I like overhang flanking the cloning site.(Fig. 3).
Fig. 3
Bsa I Not I' Spe I'
-->
5'...GG GGTCTC A^GGCC ….........^CTAG A GAGACC...3'
3'...CC CCAGAG T CCGG^TCCGGCCGCT GATC^T CTCTGG...5'
5'...GG GGTCTC A CTAG A GAGACC...3'
3'...CC CCAGAG T CCGG T CTCTGG...5'
<--
Bsa I
Please be careful Bsa 1 is not official BioBrick enzyme so you must check your plasmid backbone sequence and remove it if there is one . However you don have to worry about insert BioBricks, because they only need to be digested by official BioBrick enzymes.
For domain fusion, removal of existing stop codons of prefix and/or suffix is essential. This can be easily achieved by designing primers which delete stop codons by adding single point mutation. These primer sets can be used as universal primers which aneal to all BioBricks.
RFC submission
We have submitted this method as BBF RFC 87. For more details about RFC submission, please see also here.
 "
"