Team:Washington/Celiacs/Results
From 2011.igem.org
(→Calculating the catalytic efficiency) |
(→Calculating the catalytic efficiency) |
||
| (30 intermediate revisions not shown) | |||
| Line 48: | Line 48: | ||
[[File:Washington BestCombMutant.png|center|500px|thumb|Our final engineered enzyme showed activity over 100 fold higher than wild type Kumamolisin, and ~700 fold higher than SC-PEP.]] | [[File:Washington BestCombMutant.png|center|500px|thumb|Our final engineered enzyme showed activity over 100 fold higher than wild type Kumamolisin, and ~700 fold higher than SC-PEP.]] | ||
| + | |||
| + | |||
=='''Calculating the catalytic efficiency'''== | =='''Calculating the catalytic efficiency'''== | ||
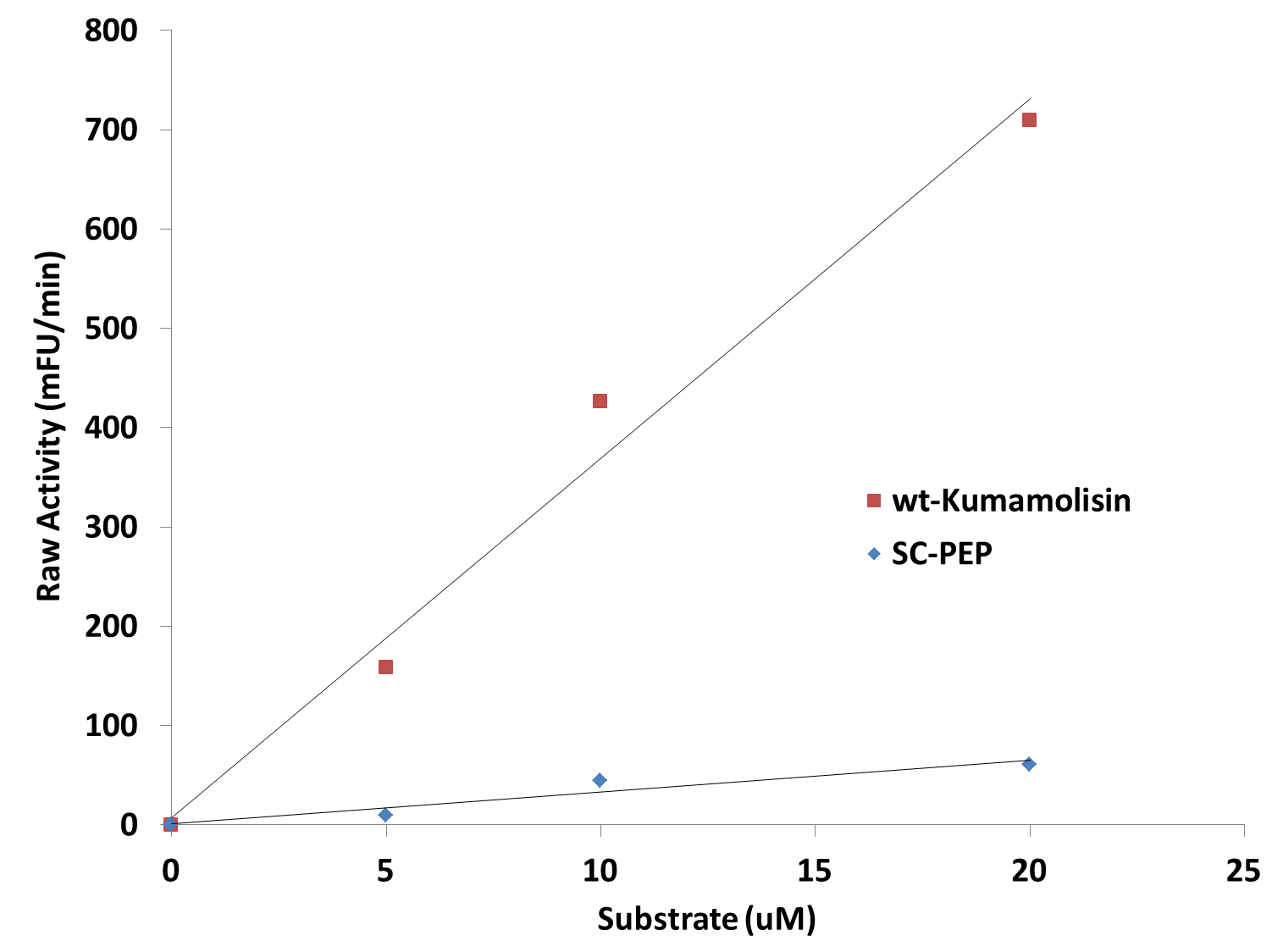
| - | [[File: | + | [[File:Washington_Linear_Michaelis_Celiac.png|right|450px|thumb|Activity vs. substrate graph shows assays were done at substrate levels where the Michaelis-Menten curve is linear. R<sup>2</sup> values were greater 0.9 for both lines of best fit.]] |
| - | Since we were able to measure the activity of these mutants at substrate and enzyme concentrations corresponding to the linear portion of a Michaelis-Menten | + | {| {{table}} |
| + | | align="center" style="background:#f0f0f0;"|'''Mutation(s)''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''''k<sub>cat</sub>''/''K<sub>M</sub>'' (M<sup>-1</sup> s<sup>-1</sup>)''' | ||
| + | |- | ||
| + | | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590023 '''N291D''']||(5.21 ± 0.11) x 10<sup>5</sup> | ||
| + | |- | ||
| + | | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590024 '''S354N, D358G, D368H''']||(2.45 ± 0.02) x 10<sup>5</sup> | ||
| + | |- | ||
| + | | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590022 '''G319S, D358G, D368H''']||(2.02 ± 0.02) x 10<sup>5</sup> | ||
| + | |- | ||
| + | | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590087 '''G319S, D358G, D368H, N291D (KumaMax)''']||(3.86 ± 0.04) x 10<sup>6</sup> | ||
| + | |- | ||
| + | | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590021 '''wt-Kumamolisin''']||(3.25 ± 0.05) x 10<sup>4</sup> | ||
| + | |- | ||
| + | | SC-PEP ||(4.96 ± 0.75) x 10<sup>3</sup> | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | Since we were able to measure the activity of these mutants at substrate and enzyme concentrations corresponding to the linear portion of a Michaelis-Menten curve, we were also able to calculate the catalytic efficiency (''k<sub>cat</sub>''/''K<sub>M</sub>'') values for wt-Kumamolisin, SC-PEP, and a few of our best mutants. This was done by first converting the raw activity in milli-Fluorescent Units (mFU)/min to the observed velocity (V<sub>obs</sub>) in M/min using a conversion constant with units of mFU/M. Since the concentration of total substrate and enzyme in each case was known, we were then able to use the equation (''k<sub>cat</sub>''/''K<sub>M</sub>'') = V<sub>obs</sub>/([E][S]), derived from the Michaelis-Menten equation, to calculate the catalytic efficiency of each mutant enzyme of interest. We were not, however, able to measure the independent catalytic (''k<sub>cat</sub>'') and binding constants (''K<sub>M</sub>'') using this methodology. Thus, one of our priorities moving forward will be to develop a mass spectroscopy assay to determine these constants independently. | ||
Latest revision as of 23:53, 28 October 2011
Testing Kumamolisin-As against SC-PEP
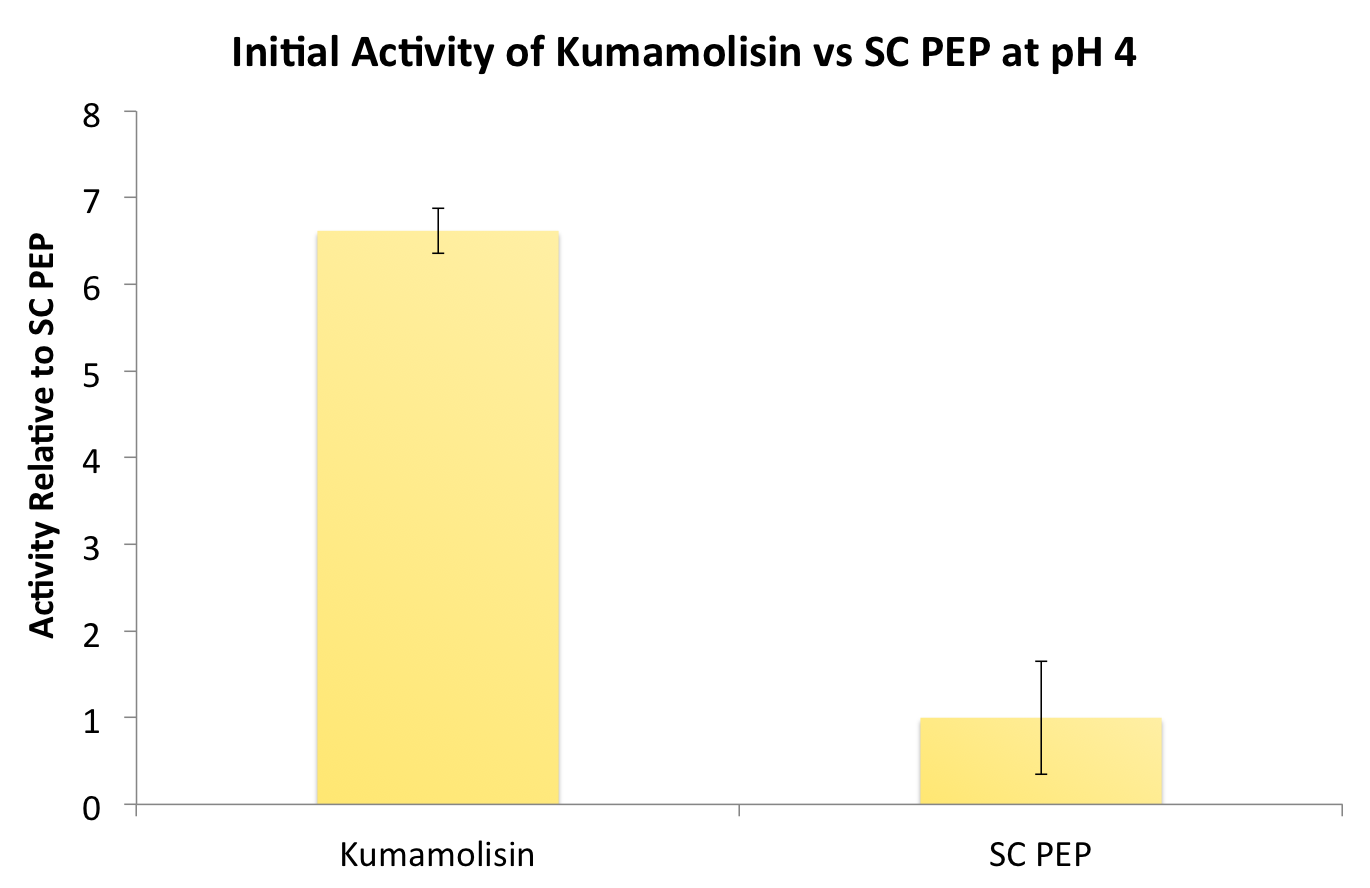
After identifying Kumamolisin as a good candidate for activity at low pH, we tested its activity on breaking down PQLP at pH 4 against the activity of SC PEP, the enzyme currently in clinical trials for breaking down gluten. Kumamolisin had never been tested for its ability to breakdown gluten, and so we began novel experimentation into the enzyme's activity on our gluten model. From tests using the fluorescent PQLP system described in our methods section, Kumamolisin showed about 7 times better activity on breaking down PQLP at pH 4 when compared to the activity of SC PEP.
Testing mutants for activity on breaking down PQLP
Using a whole cell lysate assay to screen a large number of mutants for good activity
In order to determine whether our proposed mutations to the wild-type Kumamolisin improved the ability of the enzyme to break down PQLP, we screened each mutant with a whole cell lysate fluorescence assay. Cells harboring the expressed mutants were lysed and the assay was performed at pH 4, mimicking the gastric environment. The released enzymes, after being roughly separated from cell material, were added to a fluorescent PQLP that had been conjugated to a quencher. Thus, no fluorescence was achieved until the peptide had been cleaved and the fluorophore had been released from the quencher. This allowed a relative assessment of rate of enzyme activity by measuring increase in fluorescence of the system.
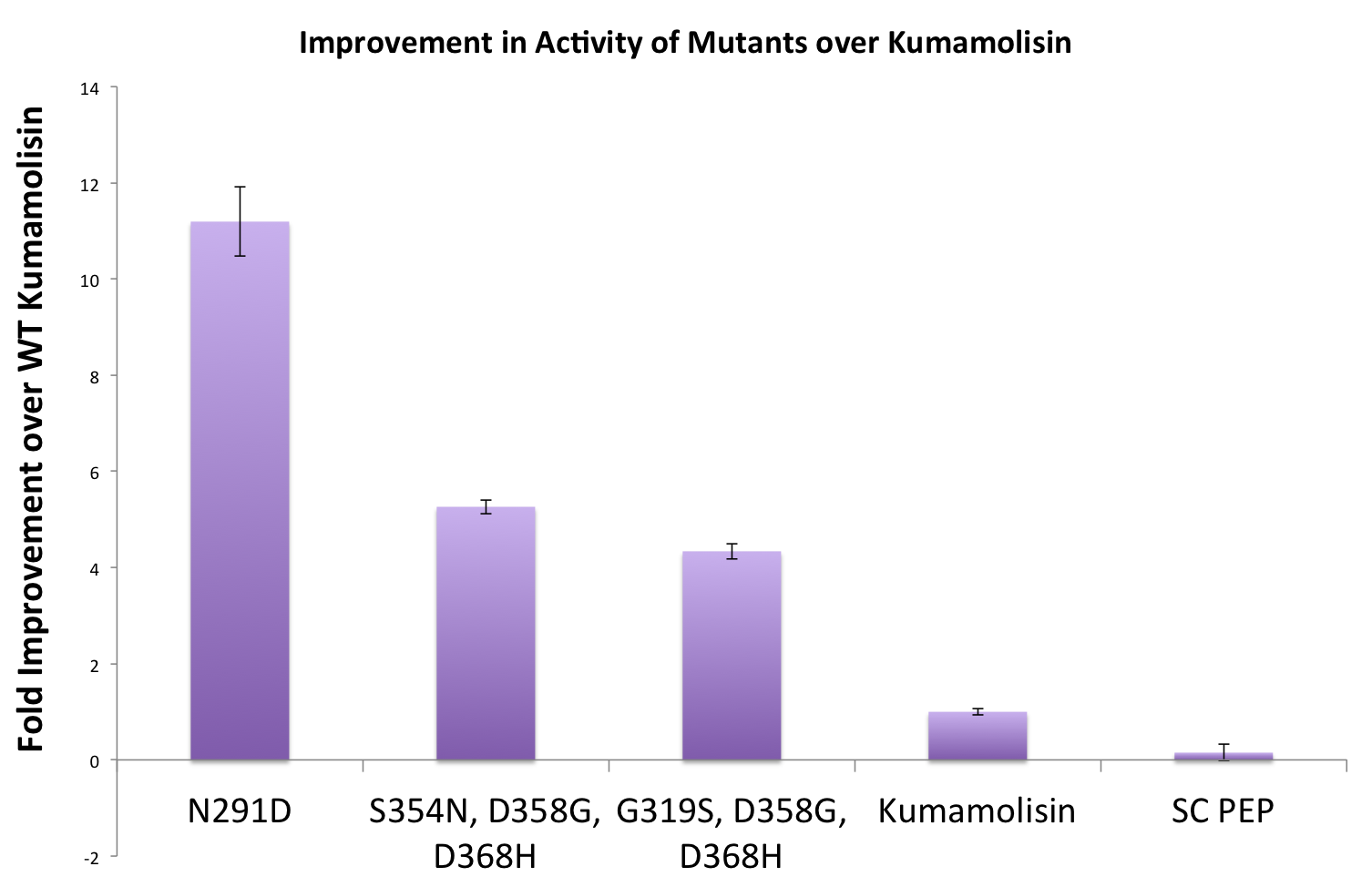
As one might expect, our first screen of mutants showed some mutants with a decrease in activity from the wild-type, some showed no change, and some actually showed great increase in activity. One single point mutant showed over 10-fold increase in activity from wild-type Kumamolisin!
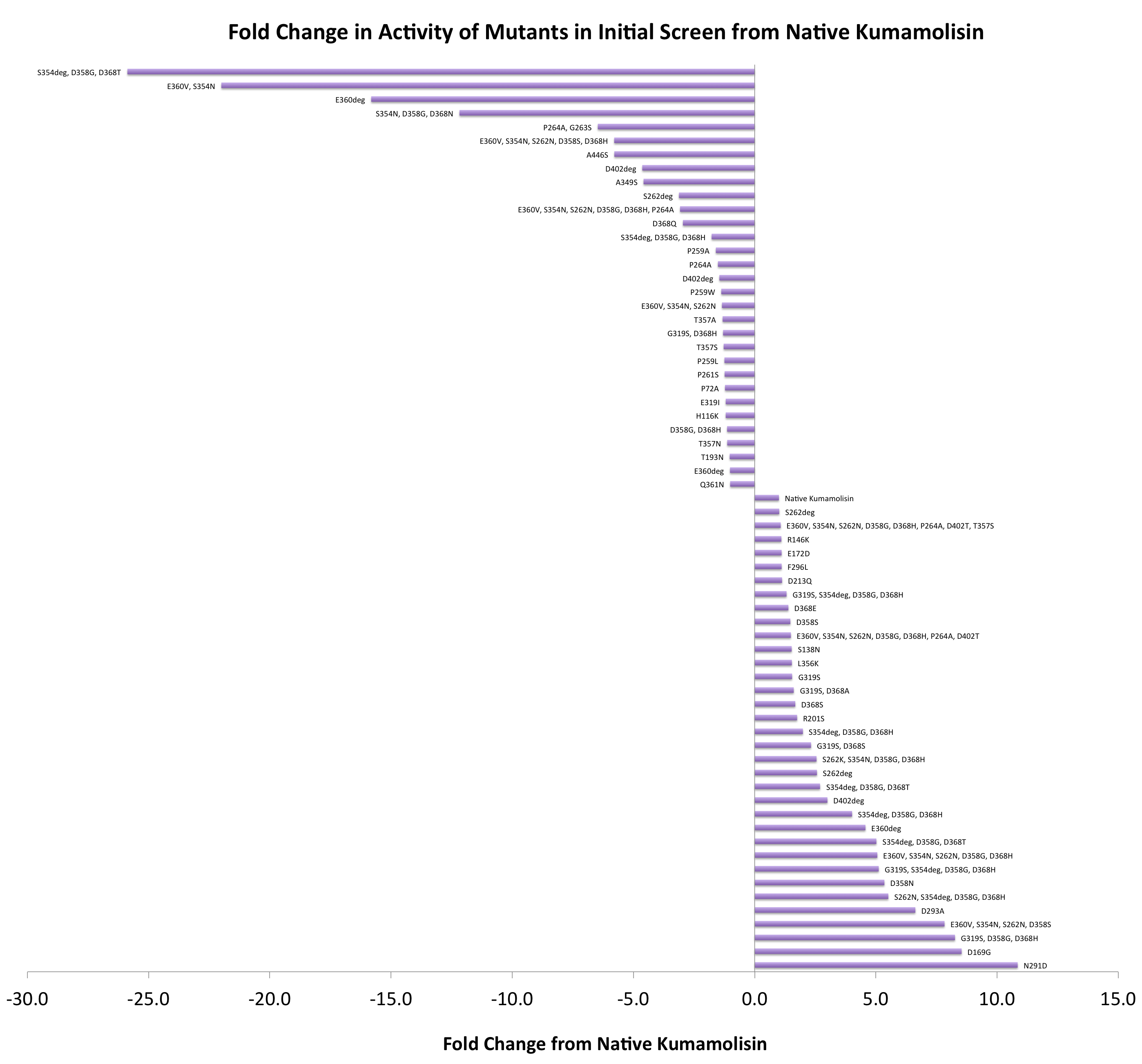
Purifying and characterizing promising mutants for accurate rate comparison
Once we had identified mutants that showed a promising increase in activity from the wild-type, we purified and characterized activity in concentration controlled fluorescence assays, identical to the fluorescence system used for the whole cell lysate assay. Our best mutant demonstrated an 11-fold increase in activity from the native enzyme.
Combining Mutations for the Construction of a Gluten Hydrolase
In order to achieve even more rate improvement from the native, we repeated our mutagenesis, this time taking successful mutations and adding them together to make combinatorial variants.
Successful mutations were combined to construct a second library for screening
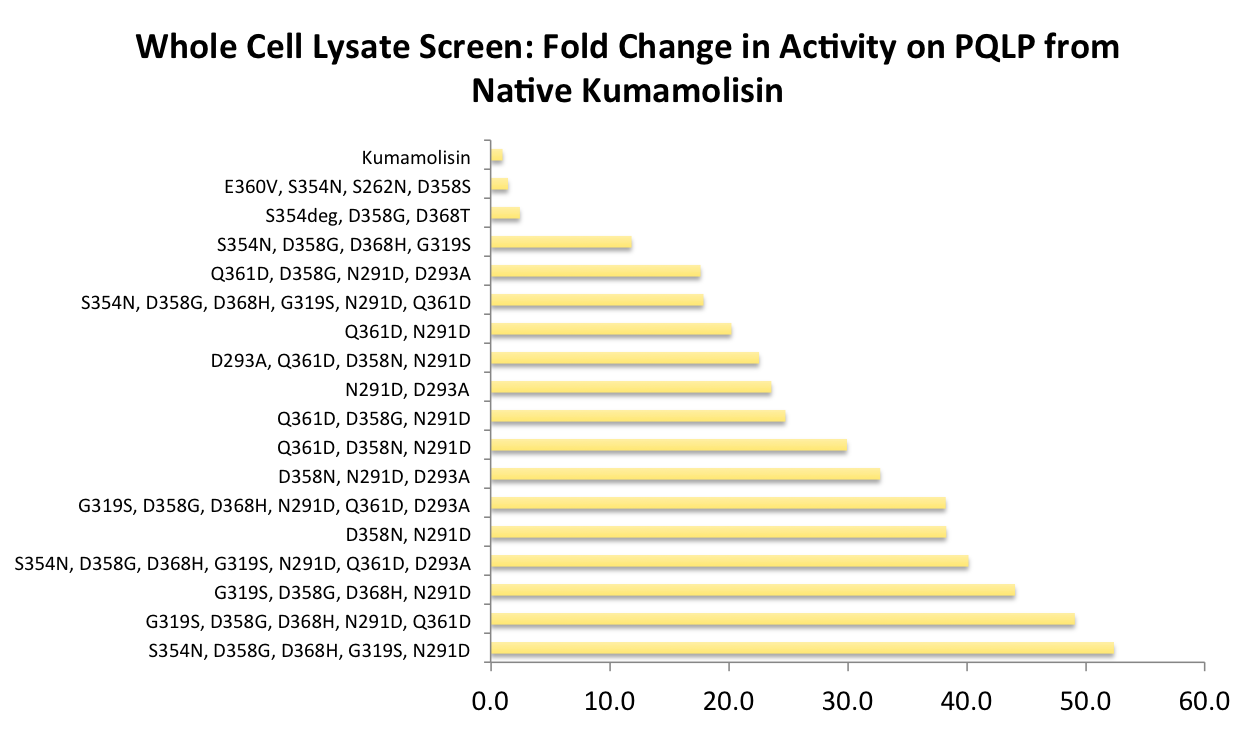
After designing a collection of combinatorial mutants, drawing from successful mutations discovered in the first round, we again performed a rough screen to identify promising combinations of mutations. From the initial screen on our combinatorial mutants, it appeared that we had achieved around 50 times better activity than native Kumamolisin on breaking down PQLP.
One of the combinatorial mutants resulted in over a 100-fold increase in activity
By combining two of our top groups of mutations from the first round, we achieved an over 100-fold increase in activity on breaking down PQLP from the wild-type enzyme. This variant enzyme is ultimately 784 times better at breaking down PQLP than SC PEP, the enzyme currently in clinical trials for treating gluten intolerance.
Calculating the catalytic efficiency
| Mutation(s) | kcat/KM (M-1 s-1) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590023 N291D] | (5.21 ± 0.11) x 105 |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590024 S354N, D358G, D368H] | (2.45 ± 0.02) x 105 |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590022 G319S, D358G, D368H] | (2.02 ± 0.02) x 105 |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590087 G319S, D358G, D368H, N291D (KumaMax)] | (3.86 ± 0.04) x 106 |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590021 wt-Kumamolisin] | (3.25 ± 0.05) x 104 |
| SC-PEP | (4.96 ± 0.75) x 103 |
Since we were able to measure the activity of these mutants at substrate and enzyme concentrations corresponding to the linear portion of a Michaelis-Menten curve, we were also able to calculate the catalytic efficiency (kcat/KM) values for wt-Kumamolisin, SC-PEP, and a few of our best mutants. This was done by first converting the raw activity in milli-Fluorescent Units (mFU)/min to the observed velocity (Vobs) in M/min using a conversion constant with units of mFU/M. Since the concentration of total substrate and enzyme in each case was known, we were then able to use the equation (kcat/KM) = Vobs/([E][S]), derived from the Michaelis-Menten equation, to calculate the catalytic efficiency of each mutant enzyme of interest. We were not, however, able to measure the independent catalytic (kcat) and binding constants (KM) using this methodology. Thus, one of our priorities moving forward will be to develop a mass spectroscopy assay to determine these constants independently.
 "
"