Team:Tsinghua/experiment
From 2011.igem.org
(→Bind & Release) |
(→Resetting) |
||
| (22 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
<div id="content_box"> | <div id="content_box"> | ||
<script> | <script> | ||
| - | navl = new Array( | + | navl = new Array("Project", "Experiments", "Modeling"); |
| - | lk = new Array(" | + | lk = new Array("/Team:Tsinghua/project", "#exp", "/Team:Tsinghua/modeling"); |
writenav(navl, lk); | writenav(navl, lk); | ||
</script> | </script> | ||
<div id="main_content"> | <div id="main_content"> | ||
| - | + | <a id="exp"></a> | |
</html> | </html> | ||
=Present proteins to the outer membrane= | =Present proteins to the outer membrane= | ||
==Molecular cloning== | ==Molecular cloning== | ||
| - | + | We fused GFP to the C-terminal of OmpA and inserted the his-tag in between them. | |
| - | + | ||
| - | + | ||
| - | + | ||
The final plasmid is as follows. | The final plasmid is as follows. | ||
| - | [[File: | + | [[File:Thuexp_og.png]][[File:Thuexp_GFPp.png]] |
==Expression Test== | ==Expression Test== | ||
| Line 252: | Line 249: | ||
=Bind & Release= | =Bind & Release= | ||
| + | |||
| + | E. coli with the plasmid for OmpA-HIV site-SH3 is induced with 0.1mM IPTG at 18 centigrade for 12 hours before harvesting by mild centrifuge. | ||
E. coli expressing OmpA-HIV protease site-SH3 was mixed with mCherry solution at room temperature for 30 minutes. | E. coli expressing OmpA-HIV protease site-SH3 was mixed with mCherry solution at room temperature for 30 minutes. | ||
| Line 257: | Line 256: | ||
After washing with distilled water twice, some of the bacteria were fixed on a slip and stained with DAPI. | After washing with distilled water twice, some of the bacteria were fixed on a slip and stained with DAPI. | ||
| - | {| style="background: none;" align="center | + | {| style="background: none; text-align:center;" align="center" |
! Color Channel | ! Color Channel | ||
! E. coli with IPTG induction | ! E. coli with IPTG induction | ||
| Line 280: | Line 279: | ||
| - | {| style="background: none;" | + | {| style="background: none; text-align:center;" align="center" |
| Color Channel | | Color Channel | ||
| + E. coli expressing protease | | + E. coli expressing protease | ||
| Line 286: | Line 285: | ||
|- | |- | ||
| mCherry | | mCherry | ||
| - | | [[File:Thuexp_release_p.png| | + | | [[File:Thuexp_release_p.png|350px]] |
| - | | [[File:Thuexp_release_n.png| | + | | [[File:Thuexp_release_n.png|350px]] |
|- | |- | ||
| DAPI | | DAPI | ||
| - | | [[File:Thuexp_release_p_d.png| | + | | [[File:Thuexp_release_p_d.png|350px]] |
| - | | [[File:Thuexp_release_n_d.png| | + | | [[File:Thuexp_release_n_d.png|350px]] |
|- | |- | ||
| merge | | merge | ||
| - | | [[File:Thuexp_release_p_m.png| | + | | [[File:Thuexp_release_p_m.png|350px]] |
| - | | [[File:Thuexp_release_n_m.png| | + | | [[File:Thuexp_release_n_m.png|350px]] |
|} | |} | ||
| + | |||
| + | =Chemotaxis= | ||
| + | |||
| + | We designed our device for testing chemotaxis. We first build a concentration gradient inside an agar bar as in any other traditional practice. When put in the water, the gradient agar bar will dictate the formation of a concentration gradient in water as solutes inside the agar can diffuse out. We use a dialysis membrane to wrap the bar and some water up, and then stir the water outside the membrane so that excess of solutes can diffuse outside the membrane and maintain the gradient inside the dialysis membrane. | ||
| + | |||
| + | [[File:Thumodel_1.png]] | ||
| + | |||
| + | ==Building agar bar== | ||
| + | |||
| + | [[File:Thuexp_bar.jpg|500px]] | ||
| + | |||
| + | An agar bar can be built stepwise to generate any gradient we need. We first tried to build an agar bar with a linear gradient of the blue dye. | ||
| + | |||
| + | [[File:Thuexp_gradbar.jpg|500px]] | ||
| + | |||
| + | ==Test of gradient in water== | ||
| + | |||
| + | We tested the formation of gradient in water by color dyes. | ||
| + | |||
| + | [[File:Thuexp_oggrad.jpg|500px]] | ||
| + | |||
| + | This color gradient maintained for over 30min, and is thus suitable for later gradient mediated chemotaxis. | ||
| + | |||
| + | ==Test of E. coli chemotaxis== | ||
| + | |||
| + | All the bacteria used here were washed with distilled water at least twice to remove traces of possible chemoattractant. | ||
| + | |||
| + | We inserted an agar bar into the capsule sealed by dialysis membrane. One end of the agar bar contained a mixture of aspartate, glutamate, serine, threonine, and cystein (10mM each). The bacteria we used was UU1250 E. coli strain (kind gift from Dr. Mark Goulian, U Penn. Δtsr-7028 Δ(tar-tap)5201 Δtrg-100 Δaer-1) harboring plasmid [http://partsregistry.org/Part:BBa_K533009 BBa_K533009], which should has only chemotaxis towards aspartate. | ||
| + | |||
| + | |||
| + | |||
| + | [[File:Thuexp_ecmov.jpg|500px]] | ||
| + | |||
| + | We expected the bacteria to form an obvious concentration difference in the capsule sealed by dialysis membrane, but no obvious difference in the cloudiness was observed in all our experiments. As our modeling put it, in a sealed space like our dialysis capsule, the cells would move towards the destination and turn around randomly, reducing the difference in cell density originally formed by chemotaxis. | ||
| + | |||
| + | Nonetheless, our mathematical model still suggests that certain difference should be present between two ends of the capsule. We took 500ul liquid from either end of the capsule and measured OD600. | ||
| + | |||
| + | As a negative control, we inserted a blank agar bar and took the same measurements. | ||
| + | |||
| + | {| style="background: none; text-align: center" align="center" | ||
| + | | style="padding: 5px; border-bottom: 1px solid #39F; border-right: 1px solid #39F;" | | ||
| + | ! style="padding: 5px 10px; border-bottom: 1px solid #39F;" | + Amino acids | ||
| + | ! style="padding: 5px 10px; border-bottom: 1px solid #39F;" | - Amino acids | ||
| + | |- | ||
| + | ! style="padding: 5px; border-right: 1px solid #39F;" | Average OD on blank end | ||
| + | | 1.47 | ||
| + | | 1.58 | ||
| + | |- | ||
| + | ! style="padding: 5px; border-right: 1px solid #39F;" | Standard Error | ||
| + | | 0.08 | ||
| + | | 0.11 | ||
| + | |- | ||
| + | ! style="padding: 5px; border-right: 1px solid #39F;" | Average OD on sweet end | ||
| + | | 1.63 | ||
| + | | 1.55 | ||
| + | |- | ||
| + | ! style="padding: 5px; border-right: 1px solid #39F; border-bottom: 1px solid #39F;" | Standard Error | ||
| + | | style="border-bottom: 1px solid #39F;"| 0.04 | ||
| + | | style="border-bottom: 1px solid #39F;"| 0.07 | ||
| + | |} | ||
| + | |||
| + | Statistically we observed a significant difference in cell density between the two ends of the capsule, confirming our hypothesis. | ||
| + | |||
| + | |||
| + | [[File:IMAG0048.jpg|300px]] [[File:IMAG0036.jpg|300px]] | ||
| + | [[File:IMAG0043.jpg|300px]] [[File: IMAG0049.jpg|300px]] | ||
| + | |||
| + | |||
| + | =Resetting= | ||
| + | |||
| + | We need to test whether the bacteria can return after reaching the end. | ||
| + | |||
| + | We incubated ''E. coli'' BW CAV8 (tar-, tsr-, tap-, trg-, aer-, cheW-, fimE-, fimB-) harboring pChemoK plasmid with or without arabinose for 1h at 30 centigrade and dipped on mineral medium supplemented with 2% glucose, 2mg/ml each of methionine, leucine, isoleucine, valine. Aspartate and serine was spread on the two ends of the semi-solid medium and incubated at 30 centigrade. | ||
| + | |||
| + | The photo was taken 30 hours after inoculation. | ||
| + | |||
| + | [[File:ThuR.png|350px]] Starting point marked by the blue line | ||
| + | |||
| + | The colony formed by ''E. coli'' without arabinose induction forms a small halo towards the aspartate end, while the colony formed by ''E. coli'' induced by arabinose almost formed no halo, and the shape is elongated along the gradient, showing some of the bacteria have changed their chemotaxis from aspartate to serine. Photos at later time points will demonstrate the chemotaxis better. | ||
| + | |||
| + | =Finally= | ||
| + | |||
| + | Thus we built this system with binding, release and transportation abilities to move any protein with multi-proline tag towards the direction dictated by the gradient. | ||
<html></div></div> | <html></div></div> | ||
Latest revision as of 18:10, 28 October 2011

Present proteins to the outer membrane
Molecular cloning
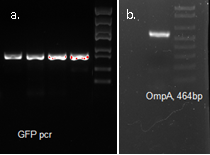
We fused GFP to the C-terminal of OmpA and inserted the his-tag in between them. The final plasmid is as follows.
Expression Test
Test for proper inducing conditions
| Group | 1 | 2 |
|---|---|---|
| Volume | 3ml | |
| IPTG(mM) | 0.1 | |
| 0.5 | ||
| 1.0 | ||
| Temperature | 30oC | 18oC |
| Duration | 12h | 12h |
| Result | Media remained yellowish. E. coli white | Media turned greenish. E. coli green |
Use 0.5mM IPTG, 18 centigrade, 12 hours induction in later experiments.
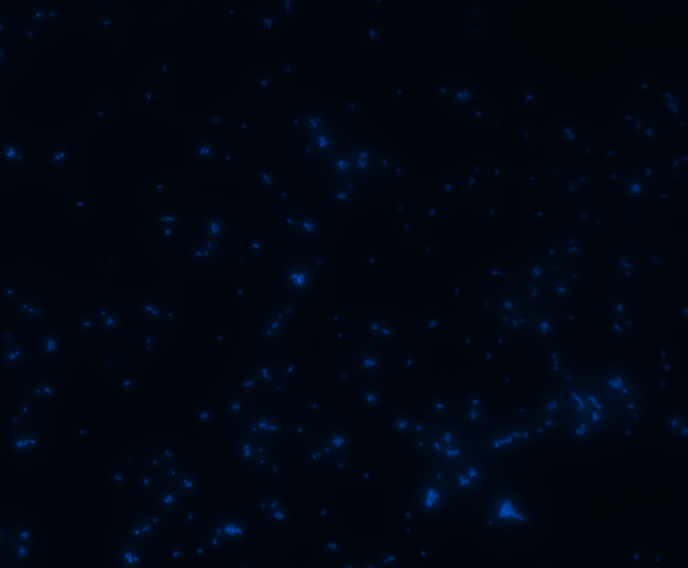
Fluorescence microscopy
Green fluorescence is bright.
In order to determine whether GFP is presented outside the membrane, cells were digested with Protease K at 50 centigrade for 30min.
| Color Channel | Post-digest | Pre-digest |
|---|---|---|
| GFP | 
| 
|
| DAPI | 
| 
|
| Merge | 
| 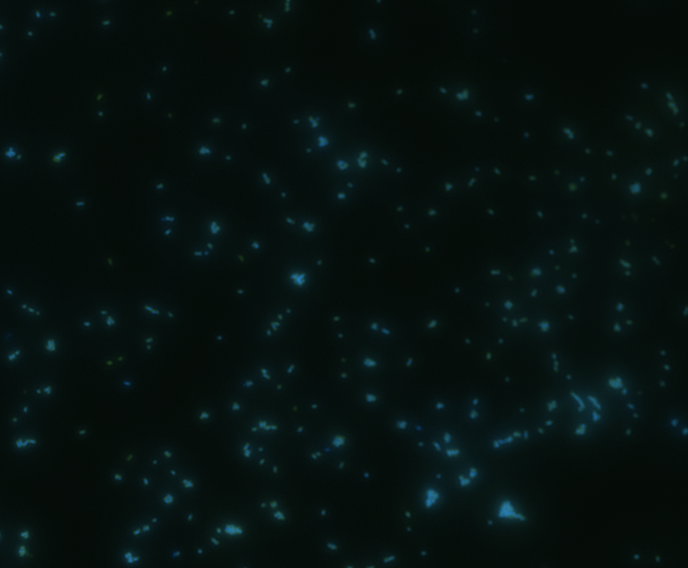
|
The fluorescence disappeared completely after digestion, which seems too good to be true. Hence, we performed Western blotting to be sure of the result.
Western Blotting
The cell components were first fractionated before blotting.
1. Sonicate the bacteria culture.
2. Centrifuge 13000rpm, 30min to separate soluble protein and membrane components from the cell debris and the non-soluble protein.
3. 51000rpm 1h to separate soluble protein from membrane components.
The majority of the OmpA-GFP protein is in the pellet after the first centrifuge, indicating incomplete cell lysis or inefficient protein folding. Nonetheless, from the later result, we can see that it is exclusively in the membrane and not in the soluble proteins.
Expression of Substrate
Molecular cloning
We synthesized the multi-proline sequence and linked it to mCherry.
Test of Expression
| Group | 1 | 2 |
|---|---|---|
| Volume | 3ml | |
| IPTG(mM) | 0.1 | |
| 0.5 | ||
| 1.0 | ||
| Temperature | 30oC | 18oC |
| Duration | 12h | 12h |
| Result | Media remained yellowish. E. coli white | Media turned purple. E. coli cherry pink |
After Pro-rich mCherry expression, significant color change can be observed (bacteria cells turned cherry red as a result of mCherry expression)
From left to right, IPTG concentration: 0, 0.1mM, 0.5mM, 1mM
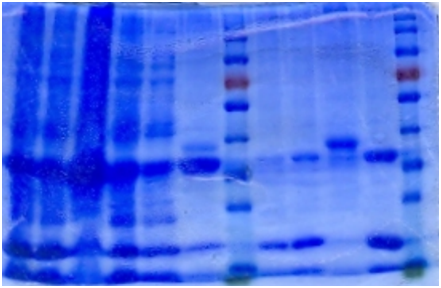
SDS-PAGE
The bacteria culture was sonicated and then centrifuged at 13000rpm for 15min. Both the supernatant and the pellet were loaded onto SDS-PAGE.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Marker | 9 | 10 | 11 | 12 | 13 | 14 |
| 1. Pellet, 1mM 18℃ | 2. Supernatant 1mM 18℃ | 3. P 0.5mM 18℃ |
| 4. S 0.5mM 18℃ | 5. P 0.1mM 18℃ | 6. S 0.1mM 18℃ |
| 7. P NC 18℃ | 8. S NC 18℃ | 9. S 1mM 30℃ |
| 10. P 1mM 30℃ | 11. S 0.5mM 30℃ | 12. P 0.5mM 30℃ |
| 13. S 0.1mM 30℃ | 14. P 0.1mM 30℃ |
Contrary to the sharp change in color, no obvious band can be seen in SDS-PAGE. Hence, further purification of protein is necessary.
Ni-column purification
The cell lysate was then purified by Ni-column. SDS-PAGE indicates that the protein is largely purified.
| 1 | 2 | 3 | 4 | 5 | 6 | Marker | 7 | 8 | 9 | 10 | Marker |
| 1. Sonicated | 2. Supernatant | 3. Pellet |
| 4. Flowthrough | 5. Buffer wash | 6. Resin |
| 7. 5mM Elution | 8. 20mM Elution | 9. Resin 2 |
| 10. 200mM Elution |
Western blotting
In order to be sure that the protein we got is multi-proline mCherry, Western blotting was performed.
As is shown, the protein eluted is indeed his-tagged mCherry.
Binding & Release Module
HIV Protease is fused with OmpA to be transported outside the membrane to facilitate substrate release.
SH3 domain is fused with OmpA as the binding module. In order to be readily released, an HIV protease site is settled in the middle.
Bind & Release
E. coli with the plasmid for OmpA-HIV site-SH3 is induced with 0.1mM IPTG at 18 centigrade for 12 hours before harvesting by mild centrifuge.
E. coli expressing OmpA-HIV protease site-SH3 was mixed with mCherry solution at room temperature for 30 minutes.
After washing with distilled water twice, some of the bacteria were fixed on a slip and stained with DAPI.
| Color Channel | E. coli with IPTG induction | E. coli without induction |
|---|---|---|
| mCherry | 
| 
|
| DAPI | 
| 
|
| merge | 
| 
|
The remaining were mixed with E. coli expressing OmpA-HIV protease at room temperature for 30 minutes to release the cargo.
After washing with distilled water twice, bacteria were fixed on a slip and stained with DAPI.
| Color Channel | + E. coli expressing protease | + E. coli without protease |
| mCherry | 
| 
|
| DAPI | 
| 
|
| merge | 
| 
|
Chemotaxis
We designed our device for testing chemotaxis. We first build a concentration gradient inside an agar bar as in any other traditional practice. When put in the water, the gradient agar bar will dictate the formation of a concentration gradient in water as solutes inside the agar can diffuse out. We use a dialysis membrane to wrap the bar and some water up, and then stir the water outside the membrane so that excess of solutes can diffuse outside the membrane and maintain the gradient inside the dialysis membrane.
Building agar bar
An agar bar can be built stepwise to generate any gradient we need. We first tried to build an agar bar with a linear gradient of the blue dye.
Test of gradient in water
We tested the formation of gradient in water by color dyes.
This color gradient maintained for over 30min, and is thus suitable for later gradient mediated chemotaxis.
Test of E. coli chemotaxis
All the bacteria used here were washed with distilled water at least twice to remove traces of possible chemoattractant.
We inserted an agar bar into the capsule sealed by dialysis membrane. One end of the agar bar contained a mixture of aspartate, glutamate, serine, threonine, and cystein (10mM each). The bacteria we used was UU1250 E. coli strain (kind gift from Dr. Mark Goulian, U Penn. Δtsr-7028 Δ(tar-tap)5201 Δtrg-100 Δaer-1) harboring plasmid [http://partsregistry.org/Part:BBa_K533009 BBa_K533009], which should has only chemotaxis towards aspartate.
We expected the bacteria to form an obvious concentration difference in the capsule sealed by dialysis membrane, but no obvious difference in the cloudiness was observed in all our experiments. As our modeling put it, in a sealed space like our dialysis capsule, the cells would move towards the destination and turn around randomly, reducing the difference in cell density originally formed by chemotaxis.
Nonetheless, our mathematical model still suggests that certain difference should be present between two ends of the capsule. We took 500ul liquid from either end of the capsule and measured OD600.
As a negative control, we inserted a blank agar bar and took the same measurements.
| + Amino acids | - Amino acids | |
|---|---|---|
| Average OD on blank end | 1.47 | 1.58 |
| Standard Error | 0.08 | 0.11 |
| Average OD on sweet end | 1.63 | 1.55 |
| Standard Error | 0.04 | 0.07 |
Statistically we observed a significant difference in cell density between the two ends of the capsule, confirming our hypothesis.
Resetting
We need to test whether the bacteria can return after reaching the end.
We incubated E. coli BW CAV8 (tar-, tsr-, tap-, trg-, aer-, cheW-, fimE-, fimB-) harboring pChemoK plasmid with or without arabinose for 1h at 30 centigrade and dipped on mineral medium supplemented with 2% glucose, 2mg/ml each of methionine, leucine, isoleucine, valine. Aspartate and serine was spread on the two ends of the semi-solid medium and incubated at 30 centigrade.
The photo was taken 30 hours after inoculation.
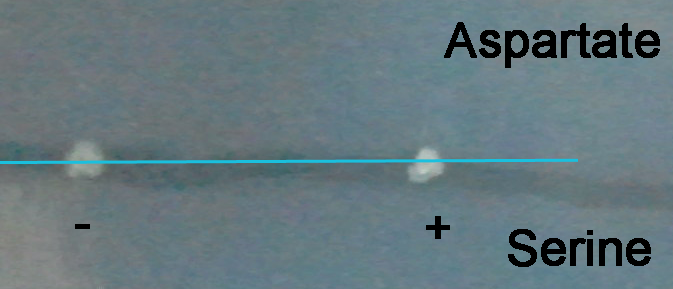 Starting point marked by the blue line
Starting point marked by the blue line
The colony formed by E. coli without arabinose induction forms a small halo towards the aspartate end, while the colony formed by E. coli induced by arabinose almost formed no halo, and the shape is elongated along the gradient, showing some of the bacteria have changed their chemotaxis from aspartate to serine. Photos at later time points will demonstrate the chemotaxis better.
Finally
Thus we built this system with binding, release and transportation abilities to move any protein with multi-proline tag towards the direction dictated by the gradient.
 "
"