Team:UCL London/Research/Supercoilometer/Experiments
From 2011.igem.org
(Created page with "{{:Team:UCL_London/Template/Header}}<html> <div id="submenu" class="sm-research-supercoilometer"> <div id="research-supercoiliology"><a href="https://2011.igem.org/Team:UCL_London...") |
|||
| (31 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<div id="research-magnetosites"><a href="https://2011.igem.org/Team:UCL_London/Research/MagnetoSites"></a></div> | <div id="research-magnetosites"><a href="https://2011.igem.org/Team:UCL_London/Research/MagnetoSites"></a></div> | ||
<div id="research-extractery"><a href="https://2011.igem.org/Team:UCL_London/Research/Extractery"></a></div> | <div id="research-extractery"><a href="https://2011.igem.org/Team:UCL_London/Research/Extractery"></a></div> | ||
| - | <div id="research- | + | <div id="research-stresslights"><a href="https://2011.igem.org/Team:UCL_London/Research/Stresslights"></a></div></html>{{:Team:UCL_London/Template/Menubar/ResearchSupercoilometer}}<html></div></html> |
<div id="content"> | <div id="content"> | ||
| - | <h1>Building a Supercoilometer in the | + | <h1>Building a Supercoilometer in the Lab</h1> |
| - | + | <h2>Aim</h2> | |
| + | To prepare a BioBrick from the tyrT promoter cloned from the E. coli genome and then construct and characterise a device which can detect the level of negative supercoiling in a closed circular plasmid | ||
| + | |||
| + | <h2>Parts used</h2> | ||
| + | <html><div align="center"><table border="1" cellspacing="0" cellpadding="3"> | ||
| + | <tr> | ||
| + | <td width="91" valign="top" bgcolor="#CCCCCC"><p><strong>Part Name</strong></p></td> | ||
| + | <td width="82" valign="top" bgcolor="#CCCCCC"><p align="center"><strong>Parts ID</strong></p></td> | ||
| + | <td width="146" valign="top" bgcolor="#CCCCCC"><p align="center"><strong>Parts Description</strong></p></td> | ||
| + | <td width="120" valign="top" bgcolor="#CCCCCC"><p align="center"><strong>Length (bp)</strong></p></td> | ||
| + | <td width="200" valign="top" bgcolor="#CCCCCC"><p align="center"><strong>Plasmid Backbone</strong></p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>RBS+CFP.LVA+Terminator</p></td> | ||
| + | <td width="82" valign="top"><p><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_B0032">BBa_E0422</a></p></td> | ||
| + | <td width="146" valign="top"><p>RBS + cyan fluorescent protein with LVA tag + terminator</p></td> | ||
| + | <td width="53" valign="top"><p align="center">917</p></td> | ||
| + | <td width="109" valign="top"><p>on pSB1A2 plasmid (2079 bp) - ampicillin resistance</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p><a href="http://partsregistry.org/Part:pSB1C3">pSB1C3</a></p></td> | ||
| + | <td width="82" valign="top"><p align="center">n/a</p></td> | ||
| + | <td width="146" valign="top"><p>standard plasmid backbone (chloramphenicol resistance)</p></td> | ||
| + | <td width="53" valign="top"><p align="center">2070</p></td> | ||
| + | <td width="109" valign="top"><p>n/a</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div></html> | ||
| + | [[File:TyrT_promoter_primers.JPG]] | ||
| + | |||
| + | ''Figure: Designed primers for PCR cloning of the tyrT promoter from E. coli genome'' | ||
| + | |||
| + | <h2>Plan for preparing the BioBricks:</h2> | ||
| + | |||
| + | # Design suitable primers for cloning out the tyrT promoter sequence from the E. coli genome and have them synthesised from Eurofins MWG Operon. The primer design will need to include the necessary BioBrick prefixes and suffixes for standardisation of the cloned part | ||
| + | # Carry out polymerase chain reaction (PCR) using the synthesised primers and Phusion polymerase to clone out the 103 bp tyrT promoter from the E. coli genome | ||
| + | # Perform purification of the PCR products and prepare them into 50 μl samples | ||
| + | # Carry out digestion of a sample of the PCR cloned tyrT promoter with EcoRI and SpeI and another sample with EcoRI and PstI | ||
| + | # Carry out digestion of linearised pSB1C3 plasmid backbone with EcoRI and PstI | ||
| + | # Transform a stock of competent TOP10 E. coli cells with the CFP expression cassette from well 8M on iGEM spring distribution - kit plate 1. Plate the cells on ampicillin plate, followed by an overnight culture in selective LB medium and finally prepare a 50 μl mini-prep sample | ||
| + | # Carry out digestion of the CFP expression cassette with XbaI and PstI | ||
| + | # Perform gel electrophoresis with a sample from the above digestion mixtures to confirm the parts | ||
| + | # Carry out ligation of the digested tyrT promoter (E/S), CFP expression cassette and the pSB1C3 plasmid backbone. Also carry out a separate ligation of the digested tyrT promoter (E/P) and the digested pSB1C3 plasmid backbone | ||
| + | # Transform separate stocks of competent TOP10 E. coli cells with the ligation mixtures and grow them overnight on chloramphenicol plates | ||
| + | # Set up overnight cultures from a single colony from the antibiotic plate in selective LB medium | ||
| + | # Set up a glycerol stock from each overnight culture (for storage at -80 °C) and mini-prep the rest to extract the plasmids with the new BioBricks for the tyrT promoter and the supercoilometer device into 50 μl samples | ||
| + | # Carry out digestion of the mini-prep samples with EcoRI and SpeI and perform gel electrophoresis to confirm the length of the new BioBricks | ||
| + | # Send a sample of the mini-prep DNA for sequencing to confirm the BioBricks | ||
| + | |||
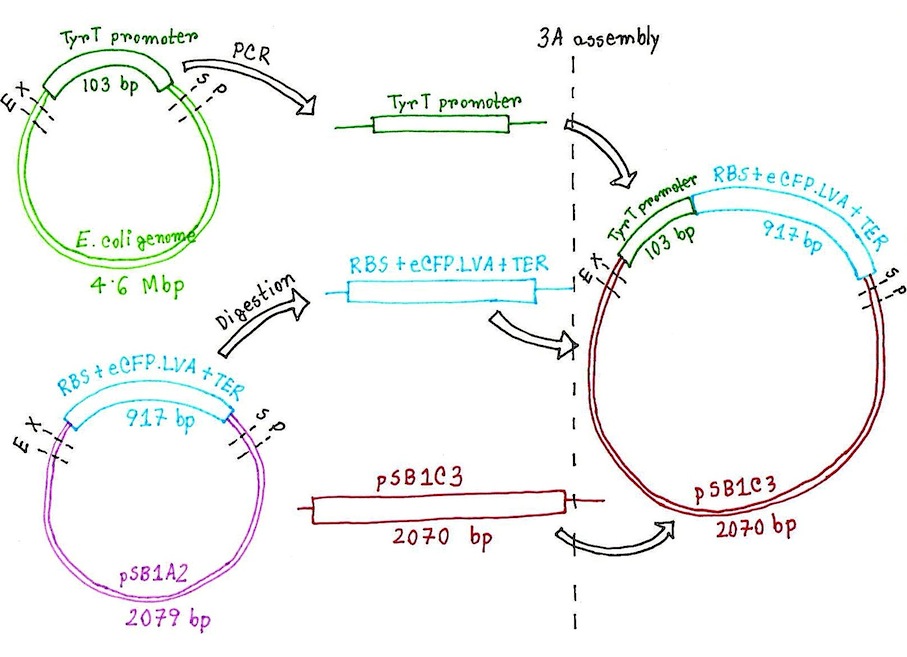
| + | [[File:Experiment_Plan_for_Supercoilometer.jpg]] | ||
| + | |||
| + | ''Figure: Overview plan for the construction of supercoilometer device.'' | ||
| + | |||
| + | <h2>Parts design submitted to the registry</h2> | ||
| + | <html><div align="center"><table border="1" cellspacing="0" cellpadding="2"> | ||
| + | <tr> | ||
| + | <td width="93" valign="top" bgcolor="#CCCCCC"><p><strong>Part Name</strong></p></td> | ||
| + | <td width="89" valign="top" bgcolor="#CCCCCC"><p align="center"><strong>Parts ID</strong></p></td> | ||
| + | <td width="150" valign="top" bgcolor="#CCCCCC"><p align="center"><strong>Parts Description</strong></p></td> | ||
| + | <td width="120" valign="top" bgcolor="#CCCCCC"><p align="center"><strong>Length (bp)</strong></p></td> | ||
| + | <td width="109" valign="top" bgcolor="#CCCCCC"><p align="center"><strong>Plasmid Backbone</strong></p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="93" valign="top"><p>tyrT promoter</p></td> | ||
| + | <td width="89" valign="top"><p><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K676003">BBa_K676003</a></p></td> | ||
| + | <td width="137" valign="top"><p>E. Coli promoter for Tyrosine tRNA operon</p></td> | ||
| + | <td width="53" valign="top"><p align="center">103</p></td> | ||
| + | <td width="200" valign="top"><p>on pSB1C3 (2070 bp) - chloramphenicol resistance</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="93" valign="top"><p>Supercoilometer</p></td> | ||
| + | <td width="89" valign="top"><p><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K676004">BBa_K676004</a></p></td> | ||
| + | <td width="137" valign="top"><p>Device to detect the level of negative supercoiling in a closed circular plasmid </p></td> | ||
| + | <td width="53" valign="top"><p align="center">1028</p></td> | ||
| + | <td width="109" valign="top"><p>on pSB1C3 (2070 bp) - chloramphenicol resistance</p></td> | ||
| + | </tr> | ||
| + | </table></div></html> | ||
| + | |||
| + | <h2>Characterisation plan for supercoilometer</h2> | ||
| + | For characterisation, it was planned to ligate the supercoilometer device to plasmid backbones containing different gyrase binding sites, previously constructed into BioBricks in our magneto-site project. The new plasmids will then be transformed into cells and cultured in shake flasks for 24 hours with regular cell samples being collected for OD readings and fluorescence assay. In this way the supercoilometer device will be placed on plasmids which will be negatively supercoiled to different extent depending on the type of GBS present and would therefore produce a difference in the level of CFP expression. A control will also need to be included in the study, using a plasmid with just the supercoilometer and no GBS. | ||
| + | |||
| + | |||
| + | Following this the level of supercoiling for each plasmid can be determined from a chloroquine gel assay, which will then provide an insight into the relationship between the extent of negative supercoiling and the quantity of CFP expression from the tyrT promoter. According to theory we expect the level of CFP fluorescence to be particularly higher for the Mu GBS/supercoilometer ligation construct, as the Mu GBS will attract gyrase strongly and introduce more negative supercoils into the plasmid then the other constructs. | ||
| + | |||
| - | |||
</div> | </div> | ||
{{:Team:UCL_London/Template/Footer}} | {{:Team:UCL_London/Template/Footer}} | ||
Latest revision as of 22:29, 7 October 2011
Contents |
Building a Supercoilometer in the Lab
Aim
To prepare a BioBrick from the tyrT promoter cloned from the E. coli genome and then construct and characterise a device which can detect the level of negative supercoiling in a closed circular plasmid
Parts used
Part Name |
Parts ID |
Parts Description |
Length (bp) |
Plasmid Backbone |
RBS+CFP.LVA+Terminator |
RBS + cyan fluorescent protein with LVA tag + terminator |
917 |
on pSB1A2 plasmid (2079 bp) - ampicillin resistance |
|
n/a |
standard plasmid backbone (chloramphenicol resistance) |
2070 |
n/a |
Figure: Designed primers for PCR cloning of the tyrT promoter from E. coli genome
Plan for preparing the BioBricks:
- Design suitable primers for cloning out the tyrT promoter sequence from the E. coli genome and have them synthesised from Eurofins MWG Operon. The primer design will need to include the necessary BioBrick prefixes and suffixes for standardisation of the cloned part
- Carry out polymerase chain reaction (PCR) using the synthesised primers and Phusion polymerase to clone out the 103 bp tyrT promoter from the E. coli genome
- Perform purification of the PCR products and prepare them into 50 μl samples
- Carry out digestion of a sample of the PCR cloned tyrT promoter with EcoRI and SpeI and another sample with EcoRI and PstI
- Carry out digestion of linearised pSB1C3 plasmid backbone with EcoRI and PstI
- Transform a stock of competent TOP10 E. coli cells with the CFP expression cassette from well 8M on iGEM spring distribution - kit plate 1. Plate the cells on ampicillin plate, followed by an overnight culture in selective LB medium and finally prepare a 50 μl mini-prep sample
- Carry out digestion of the CFP expression cassette with XbaI and PstI
- Perform gel electrophoresis with a sample from the above digestion mixtures to confirm the parts
- Carry out ligation of the digested tyrT promoter (E/S), CFP expression cassette and the pSB1C3 plasmid backbone. Also carry out a separate ligation of the digested tyrT promoter (E/P) and the digested pSB1C3 plasmid backbone
- Transform separate stocks of competent TOP10 E. coli cells with the ligation mixtures and grow them overnight on chloramphenicol plates
- Set up overnight cultures from a single colony from the antibiotic plate in selective LB medium
- Set up a glycerol stock from each overnight culture (for storage at -80 °C) and mini-prep the rest to extract the plasmids with the new BioBricks for the tyrT promoter and the supercoilometer device into 50 μl samples
- Carry out digestion of the mini-prep samples with EcoRI and SpeI and perform gel electrophoresis to confirm the length of the new BioBricks
- Send a sample of the mini-prep DNA for sequencing to confirm the BioBricks
Figure: Overview plan for the construction of supercoilometer device.
Parts design submitted to the registry
Part Name |
Parts ID |
Parts Description |
Length (bp) |
Plasmid Backbone |
tyrT promoter |
E. Coli promoter for Tyrosine tRNA operon |
103 |
on pSB1C3 (2070 bp) - chloramphenicol resistance |
|
Supercoilometer |
Device to detect the level of negative supercoiling in a closed circular plasmid |
1028 |
on pSB1C3 (2070 bp) - chloramphenicol resistance |
Characterisation plan for supercoilometer
For characterisation, it was planned to ligate the supercoilometer device to plasmid backbones containing different gyrase binding sites, previously constructed into BioBricks in our magneto-site project. The new plasmids will then be transformed into cells and cultured in shake flasks for 24 hours with regular cell samples being collected for OD readings and fluorescence assay. In this way the supercoilometer device will be placed on plasmids which will be negatively supercoiled to different extent depending on the type of GBS present and would therefore produce a difference in the level of CFP expression. A control will also need to be included in the study, using a plasmid with just the supercoilometer and no GBS.
Following this the level of supercoiling for each plasmid can be determined from a chloroquine gel assay, which will then provide an insight into the relationship between the extent of negative supercoiling and the quantity of CFP expression from the tyrT promoter. According to theory we expect the level of CFP fluorescence to be particularly higher for the Mu GBS/supercoilometer ligation construct, as the Mu GBS will attract gyrase strongly and introduce more negative supercoils into the plasmid then the other constructs.
 "
"