Team:Wageningen UR/Project/CompleteProject1Description
From 2011.igem.org
(→Synchroscillator) |
(→Synchroscillator) |
||
| (11 intermediate revisions not shown) | |||
| Line 41: | Line 41: | ||
There are a number of genetic circuit topologies that have the potential to exhibit oscillatory behavior under the right conditions. However, the requirement that the oscillations should be synchronized posed a constraint on the components that could be used. The starting point for our genetic circuitry was a design recently published in the article “A synchronized quorum of genetic clocks” by Danino et al. This design combines elements of the ''Vibrio fischeri'' quorum sensing system with a quorum quenching enzyme from ''Bacillus subtilis'', resulting in coupled positive and negative feedback loops which regulate the expression of a reporter protein. | There are a number of genetic circuit topologies that have the potential to exhibit oscillatory behavior under the right conditions. However, the requirement that the oscillations should be synchronized posed a constraint on the components that could be used. The starting point for our genetic circuitry was a design recently published in the article “A synchronized quorum of genetic clocks” by Danino et al. This design combines elements of the ''Vibrio fischeri'' quorum sensing system with a quorum quenching enzyme from ''Bacillus subtilis'', resulting in coupled positive and negative feedback loops which regulate the expression of a reporter protein. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
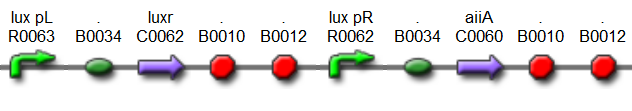
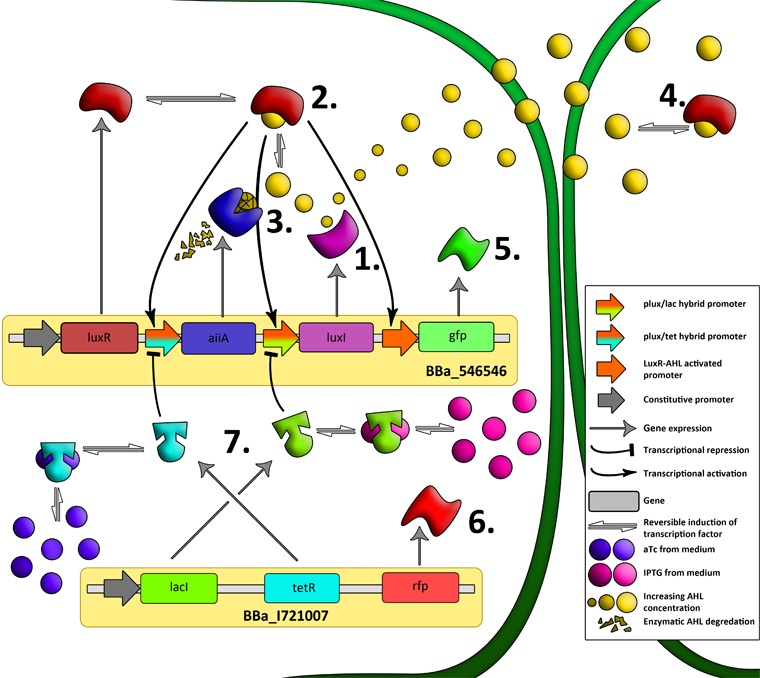
'''Basic Components:''' | '''Basic Components:''' | ||
| Line 56: | Line 50: | ||
| - | '''AiiA''' is an enzyme from B. subtilis which degrades AHL. Its biological function is to interfere with the quorum sensing signals of other bacteria. Placing it under control of the pRight promoter results in negative feedback as a response to increasing AHL concentrations. | + | '''AiiA''' is an enzyme from ''B. subtilis'' which degrades AHL. Its biological function is to interfere with the quorum sensing signals of other bacteria. Placing it under control of the pRight promoter results in negative feedback as a response to increasing AHL concentrations. |
| Line 63: | Line 57: | ||
| + | |||
| + | |||
| + | [[File:System1.png|760px|link=https://static.igem.org/mediawiki/2011/d/d0/System1.png]] | ||
| + | |||
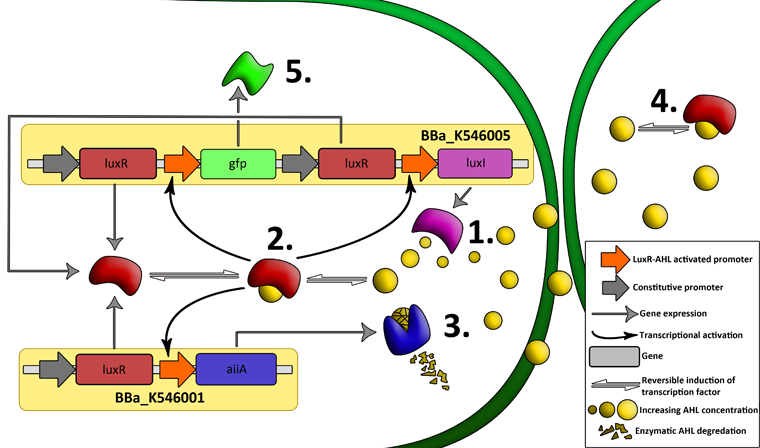
| + | '''Fig.2.''' ''Genetic circuit of a synchronized oscillator on 2 plasmids'' | ||
| + | |||
| + | '''1''': The AHL Synthase LuxI is expressed at a basal level when AHL concentration is low. Relative AHL concentration increases as a function of cell density. | ||
| + | |||
| + | '''2''': As the concentration of AHL increases, it associates with the constitutively expressed transcription factor LuxR, thereby up-regulating the expression of AiiA, LuxI and GFP. As LuxI accumulates, the concentration of AHL increases rapidly as a result of the LuxR-AHL -> LuxI positive feedback loop. | ||
| + | |||
| + | '''3''': The AHL-degrading enzyme AiiA does not accumulate as rapidly as LuxI. However, the degradation of AHL occurs at a faster rate than AHL synthesis, effectively reducing the level of active LuxR (and its transcriptional activity). This delayed negative feedback results in oscillations in AHL concentration. | ||
| + | |||
| + | '''4''': AHL diffuses freely between cells, synchronizing LuxR-AHL dependent transcriptional activity across a population of cells. | ||
| + | |||
| + | '''5''': LuxR-AHL dependent GFP expression allows the fluctuations in AHL concentration to be observed in situ. LuxI, AiiA and GFP all have LVA tags and are degraded rapidly (albeit at slightly different rates). | ||
[[Team:Wageningen_UR/Project/CompleteProject1Description#Project Description| back to top]] | [[Team:Wageningen_UR/Project/CompleteProject1Description#Project Description| back to top]] | ||
| Line 96: | Line 105: | ||
[[File:System2.png|760px|link=https://static.igem.org/mediawiki/2011/a/a8/System2.png]] | [[File:System2.png|760px|link=https://static.igem.org/mediawiki/2011/a/a8/System2.png]] | ||
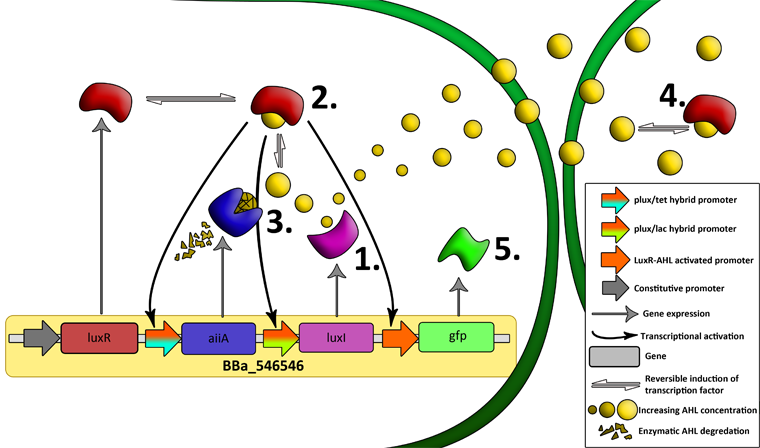
| - | '''Fig.6.''' ''Genetic circuit of | + | '''Fig.6.''' ''Genetic circuit of the synchronized oscillating system on a single plasmid.'' |
| Line 103: | Line 112: | ||
'''Fig.7.''' ''Genetic circuit of a tunable synchronized oscillator.'' | '''Fig.7.''' ''Genetic circuit of a tunable synchronized oscillator.'' | ||
| + | |||
| + | '''1''': The AHL Synthase LuxI is expressed at a basal level when AHL concentration is low. Relative AHL concentration increases as a function of cell density. | ||
| + | |||
| + | '''2''': As the concentration of AHL increases, it associates with the constitutively expressed transcription factor LuxR, thereby up-regulating the expression of AiiA, LuxI and GFP. As LuxI accumulates, the concentration of AHL increases rapidly as a result of the LuxR-AHL -> LuxI positive feedback loop. | ||
| + | |||
| + | '''3''': The AHL-degrading enzyme AiiA does not accumulate as rapidly as LuxI. However, the degradation of AHL occurs at a faster rate than AHL synthesis, effectively reducing the level of active LuxR (and its transcriptional activity). This delayed negative feedback results in oscillations in AHL concentration. | ||
| + | |||
| + | '''4''': AHL diffuses freely between cells, synchronizing LuxR-AHL dependent transcriptional activity across a population of cells. | ||
| + | |||
| + | '''5''': LuxR-AHL dependent GFP expression allows the fluctuations in AHL concentration to be observed in situ. LuxI, AiiA and GFP all have LVA tags and are degraded rapidly (albeit at slightly different rates). | ||
| + | |||
| + | '''6''': Constitutively expressed RFP allows the distinction between the effects of cell growth and of the true oscillations of GFP expression due to our system. | ||
| + | |||
| + | '''7''': The repressors LacI and TetR are constitutively expressed. They inhibit the expression of luxI and aiiA respectively. Adding IPTG removes the LacI repressor and aTc removes the TetR repressor. | ||
[[File:Dt.png|x67px|link=http://partsregistry.org/Part:BBa_K546546]] | [[File:Dt.png|x67px|link=http://partsregistry.org/Part:BBa_K546546]] | ||
| Line 164: | Line 187: | ||
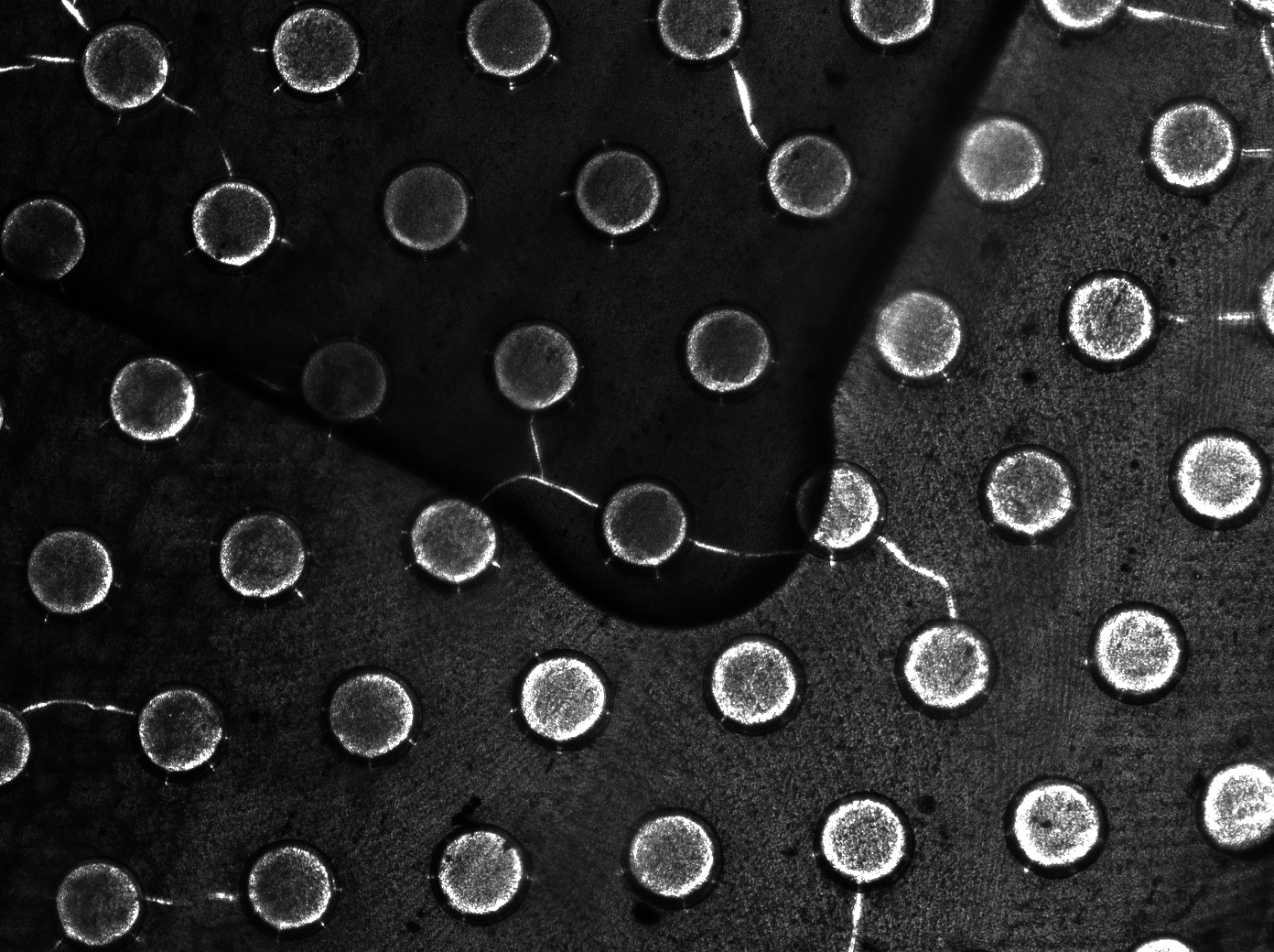
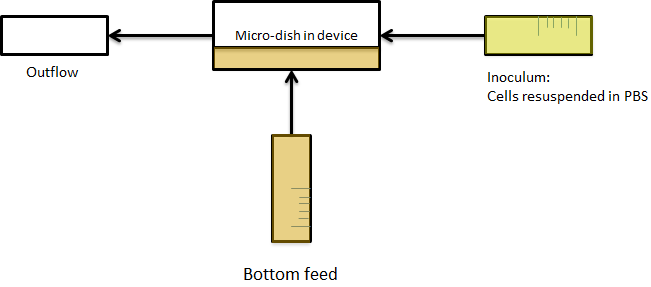
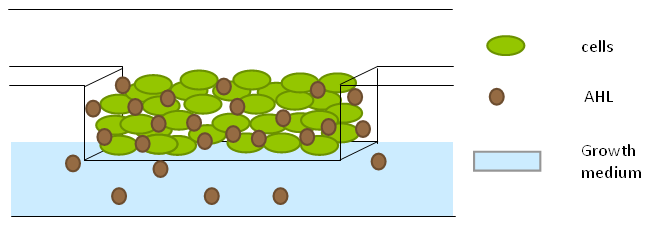
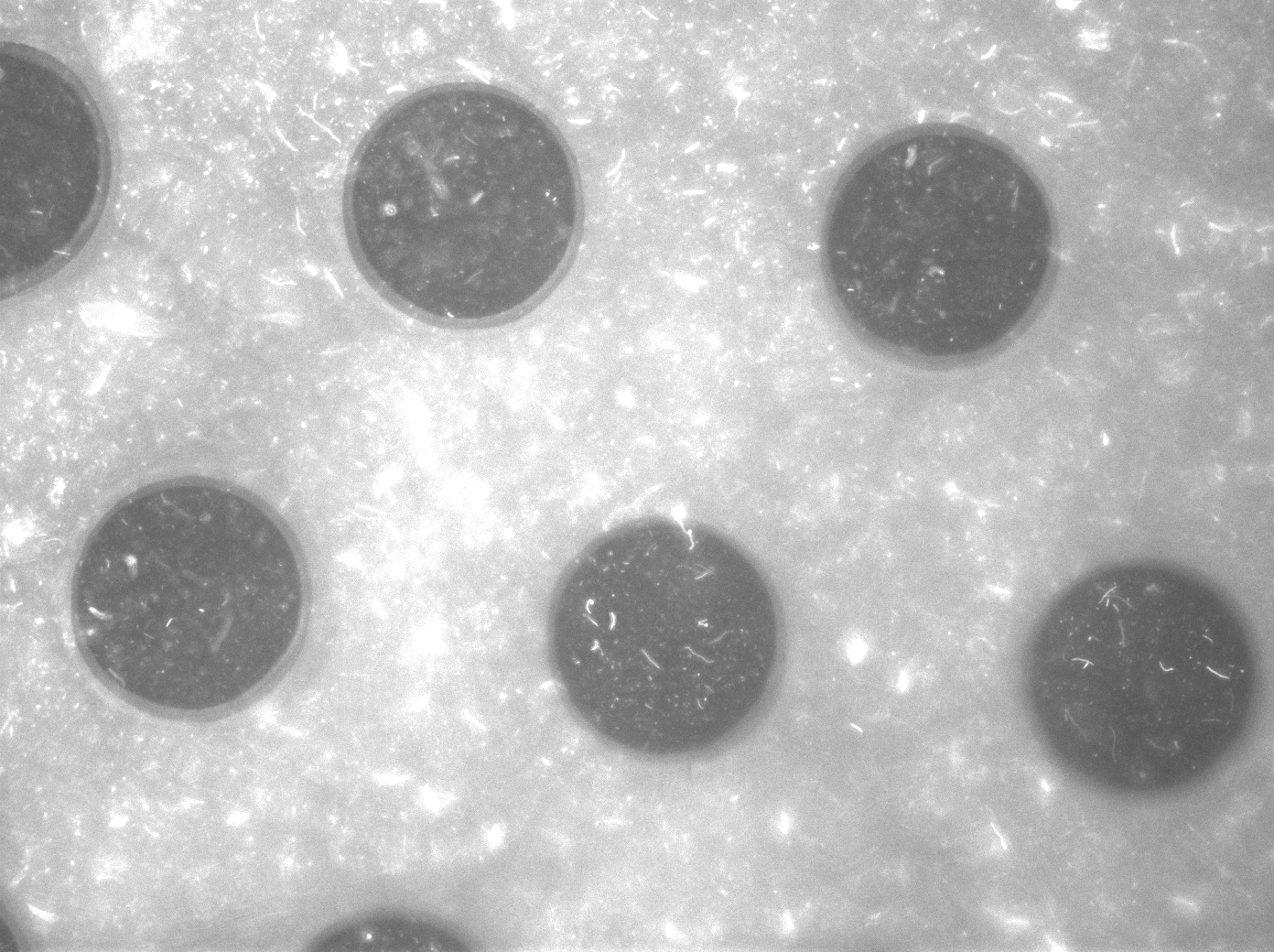
Per run four wells of the microdish were analyzed, as this is the maximal number of wells that one can focus on with a 10x objective. Pictures were processed with [http://rsbweb.nih.gov/ij/ ImageJ], a program that can measure the different wells separately. | Per run four wells of the microdish were analyzed, as this is the maximal number of wells that one can focus on with a 10x objective. Pictures were processed with [http://rsbweb.nih.gov/ij/ ImageJ], a program that can measure the different wells separately. | ||
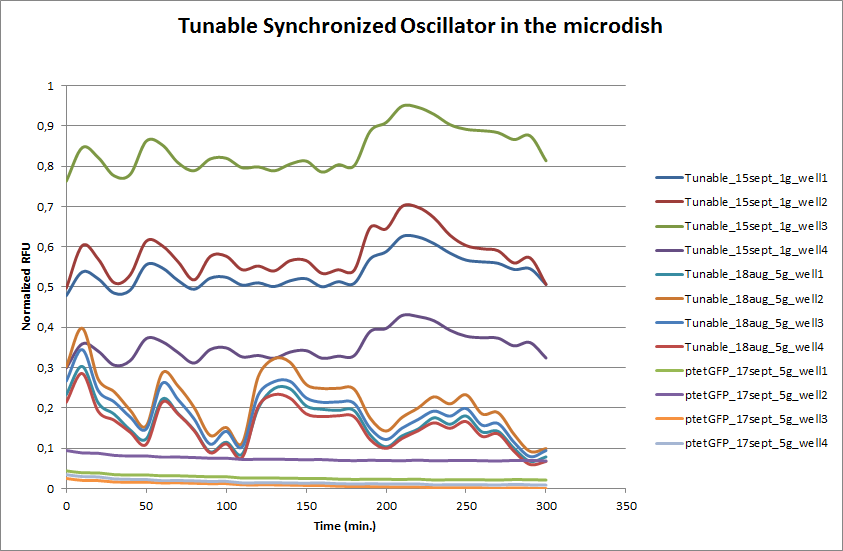
| - | These time-lapse microscopy studies of cells harboring the Synchroscillator plasmid consistently showed significant periodic changes in GFP expression compared to control experiments with cells constitutively expressing GFP (Fig.15). The observed periodicity is in the range of 60 minutes, and the asymmetry of the peaks is indicative of differences between GFP synthesis and degradation. The oscillations were sustained over multiple periods for a duration of 5 hours | + | These time-lapse microscopy studies of cells harboring the Synchroscillator plasmid consistently showed significant periodic changes in GFP expression compared to control experiments with cells constitutively expressing GFP (Fig.15). The observed periodicity is in the range of 60 minutes, and the asymmetry of the peaks is indicative of differences between GFP synthesis and degradation. The oscillations were sustained over multiple periods for a duration of 5 hours after which the expression level of GFP decreased; most likely due to decreases in cell vitality. These findings are consistent with predictions made from mathematical simulations as well as previously published results. It is noteworthy that these dynamics were able to emerge under zero-flow conditions and did not require manipulation the external AHL degradation rate. The synchronization between wells suggests that the diffusion of extracellular AHL through the bottom of the microdish occurred at a rate that allowed spatially separated cell populations to experience similar AHL concentrations. |
| Line 173: | Line 196: | ||
<html><iframe width="420" height="315" src="http://www.youtube.com/embed/Q6tDQNIaklg" frameborder="0" allowfullscreen></iframe></html> | <html><iframe width="420" height="315" src="http://www.youtube.com/embed/Q6tDQNIaklg" frameborder="0" allowfullscreen></iframe></html> | ||
| - | '''Fig.15''' '' | + | '''Fig.15''' ''Time-lapse video of the Tunable Oscillator.'' |
====6. Conclusion and Future work==== | ====6. Conclusion and Future work==== | ||
| + | |||
| + | Over the course of our experiments we were able to show that quorum sensing genes and regulatory elements composed to form interconnected positive and negative feedback loops can result in sustained synchronized oscillations under appropriate experimental conditions. | ||
| + | |||
| + | These findings highlight the utility of custom designed micro-environments for the study of complex genetic circuits. Our flow chamber outfitted with the microdish proved to be a suitable platform to investigate the temporal dynamics of intercellular communication. The ability to manipulate extracellular conditions without disrupting localized cell populations should prove useful in investigating biological systems in real time. | ||
| + | |||
| + | Future work should focus on defining appropriate wet-lab experiments in order to enhance the quality of the mathematical model by fitting it with the right parameters. Also the stability of the oscillations can be improved by optimizing the supply of nutrients to the bacteria in the wells in order to achieve long term viability of the cell cultures. Additionally, the tunability of the system could enhance the robustness of such synthetic regulatory systems. Modularity BioBrick parts expand functionality without having to redesign the system from the ground up. | ||
| + | |||
| + | ===Links and references=== | ||
| + | |||
| + | [1][http://www.nature.com/nature/journal/v463/n7279/abs/nature08753.html Danino et al. 2010] | ||
| + | |||
| + | [2][http://www.nature.com/nature/journal/v463/n7279/suppinfo/nature08753.html Supplementary information] | ||
| + | |||
| + | [3][https://2007.igem.org/Tokyo/Works Tokyo iGEM 2007] | ||
| + | |||
| + | [4][http://rsif.royalsocietypublishing.org/content/early/2010/06/28/rsif.2010.0183.short A comparative analysis of synthetic genetic oscillators] | ||
| + | |||
Latest revision as of 03:53, 22 September 2011
 "
"