Team:Edinburgh/Artificial Selection
From 2011.igem.org
| (46 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html><script type="text/javascript" >$(document).ready(function() { | <html><script type="text/javascript" >$(document).ready(function() { | ||
getMenus('model', 'model_artificial_selection'); | getMenus('model', 'model_artificial_selection'); | ||
| - | |||
}); </script></html> | }); </script></html> | ||
<div class="main_body"> | <div class="main_body"> | ||
| - | + | <p class="h1">Artificial Selection</p> | |
| - | + | As part of our feasibility study, we decided to investigate the possibility that expressing <span class="hardword" id="cellulase">cellulases</span> on the surface of ''E. coli'' might make it easier to simply evolve a system that has improved ability to degrade <span class="hardword" id="cellulose">cellulose</span>. | |
| + | |||
| + | ==Explanation== | ||
| + | |||
| + | Suppose, like most people, you want to create <span class="hardword" id="ec">''E. coli''</span> that is really efficient at degrading cellulose. Rather than do a bunch of difficult genetic engineering work, why not let Darwin do the work for you? If you simply get your ''E. coli'' working at 1% efficiency, can't you just use <span class="hardword" id="artificialselection">artificial selection</span> to get you the remaining 99% of the way? | ||
| + | |||
| + | If cellulose is available as a food source for your culture, then it should work: the cells will only get food if they can break down cellulose, and so there is a <span class="hardword" id="selectionpressure">selection pressure</span> to get better at doing so. But there's a problem noted by [http://www.springerlink.com/content/4l4m28lp06120253/ Van Zyl ''et al'' (2007)]: if your ''E. coli'' simply secrete cellulases into the media, then a rare mutant that produces more or better cellulases will not gain much benefit, since these cellulases float away and will probably benefit other cells instead. | ||
However, if the cellulases are attached to the cell '''(for example, because they are part of a fusion to Ice Nucleation Protein...)''' then this problem does not arise; the rare mutant gains all of the benefit, since the sugar that is created is right there beside it. | However, if the cellulases are attached to the cell '''(for example, because they are part of a fusion to Ice Nucleation Protein...)''' then this problem does not arise; the rare mutant gains all of the benefit, since the sugar that is created is right there beside it. | ||
| - | To demonstrate this, enter | + | To demonstrate this, enter Edinburgh's ''Celluvolve'' computer simulation! |
| + | |||
| + | ==Rules== | ||
'''The rules are as follows:''' | '''The rules are as follows:''' | ||
| - | * | + | * There is an array; every point contains a bacterial cell. |
| - | * Each | + | * Each cell has a "genotype": its ability to produce cellulase. |
* The genotype is a number between 0 and 255. | * The genotype is a number between 0 and 255. | ||
| - | * Each iteration, some random bacteria are | + | * Every iteration, every cell produces that many cellulase molecules. |
| - | * | + | * When produced, a cellulase molecule either: |
| - | * Food supply is defined | + | ** stays where it was produced [LEFT SIMULATION] or |
| - | + | ** floats a short distance randomly [RIGHT SIMULATION] | |
| - | + | * Each iteration, some random bacteria are given a chance to reproduce. | |
| - | * Reproduction means: | + | * Cells with higher food supply have a greater chance to reproduce. |
| - | + | * Food supply is defined as the amount of cellulase present at the cell. | |
| - | + | * Reproduction means: | |
| - | + | ** Overwriting a nearby cell with one's own genotype. | |
| + | ** Reproduction is not exact. There is "mutation". | ||
| + | ** The offspring's genotype is a number close to the parent's. | ||
| - | The left half of the simulation models the case where the cellulase | + | The left half of the simulation models the case where the cellulase produced by a cell can only benefit itself. The right half models the case where the cellulase can float away and benefit neighbours instead of the cell that produced it. |
| - | produced by a cell can only benefit itself. The right half models the | + | |
| - | case where the cellulase can float away and benefit neighbours instead | + | |
| - | of the cell that produced it. | + | |
| - | == | + | ==C code== |
| - | + | C code for the simulator is [https://static.igem.org/mediawiki/2011/b/bb/Celluvolve.txt available]. Note that, by default, it will spit out about 62 MB of .bmp graphics files so you can see visually what is going on. | |
| + | |||
| + | A much faster, but less mathematically rigorous version, is [https://static.igem.org/mediawiki/2011/e/e8/Celluvolve-fast.txt also available]. | ||
==Simulation run== | ==Simulation run== | ||
| - | In this simulation, each pixel represents a bacterium. Its brightness indicates its ability to produce cellulase. In the left half of the simulation, the cellulase | + | In this simulation, each pixel represents a bacterium. Its brightness indicates its ability to produce cellulase. In the left half of the simulation, the amount of cellulase available to a cell is entirely dependent on its genotype, whereas in the right half, the amount also depends on the genotype of nearby cells. |
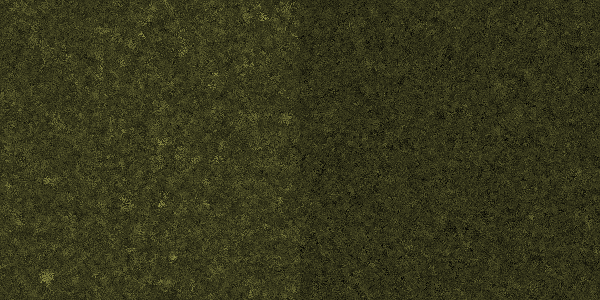
| - | [[File: | + | [[File:Celluvolve3-200.png|center|thumb|600px|caption|Generation 200: Hardly a noticable difference...]] |
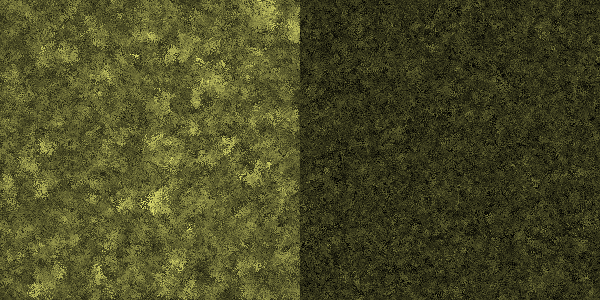
| - | [[File: | + | [[File:Celluvolve3-400.png|center|thumb|600px|caption|Generation 400: Evolution is making more of a difference on the left side...]] |
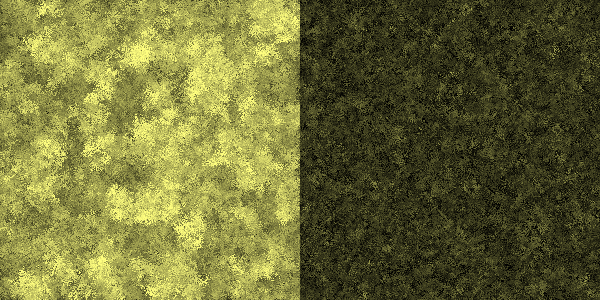
| - | [[File: | + | [[File:Celluvolve3-600.png|center|thumb|600px|caption|Generation 600: Foom... bacteria on the left side are now much better at degrading cellulose. The right side has improved just a little.]] |
==Graph== | ==Graph== | ||
| - | [[File: | + | Here's a graph of the simulation... |
| + | |||
| + | |||
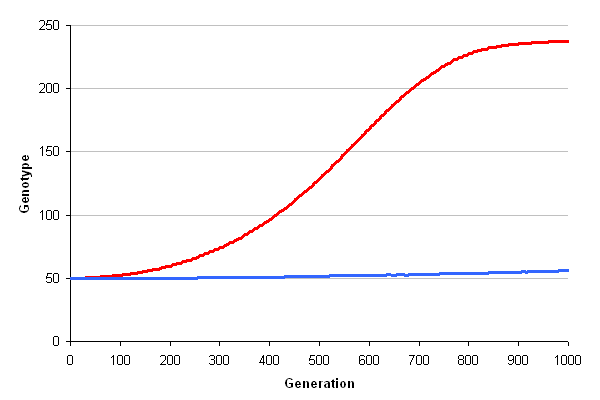
| + | [[File:Celluvolve3-graph.png|center|thumb|600px|caption|Graph of average genotype (i.e. ability to make cellulase) over time. Red is the case where cellulases attach to the cell producing them; blue is the case where cellulases can float away.]] | ||
| + | |||
| + | ==Conclusion== | ||
| + | |||
| + | This is an excellent reason why surface display of cellulases is preferable to releasing them into the media. Surface display drastically improves the prospects for simply evolving the desired phenotype. | ||
==References== | ==References== | ||
Latest revision as of 14:41, 10 September 2011
Artificial Selection
As part of our feasibility study, we decided to investigate the possibility that expressing cellulases on the surface of E. coli might make it easier to simply evolve a system that has improved ability to degrade cellulose.
Contents |
Explanation
Suppose, like most people, you want to create E. coli that is really efficient at degrading cellulose. Rather than do a bunch of difficult genetic engineering work, why not let Darwin do the work for you? If you simply get your E. coli working at 1% efficiency, can't you just use artificial selection to get you the remaining 99% of the way?
If cellulose is available as a food source for your culture, then it should work: the cells will only get food if they can break down cellulose, and so there is a selection pressure to get better at doing so. But there's a problem noted by [http://www.springerlink.com/content/4l4m28lp06120253/ Van Zyl et al (2007)]: if your E. coli simply secrete cellulases into the media, then a rare mutant that produces more or better cellulases will not gain much benefit, since these cellulases float away and will probably benefit other cells instead.
However, if the cellulases are attached to the cell (for example, because they are part of a fusion to Ice Nucleation Protein...) then this problem does not arise; the rare mutant gains all of the benefit, since the sugar that is created is right there beside it.
To demonstrate this, enter Edinburgh's Celluvolve computer simulation!
Rules
The rules are as follows:
- There is an array; every point contains a bacterial cell.
- Each cell has a "genotype": its ability to produce cellulase.
- The genotype is a number between 0 and 255.
- Every iteration, every cell produces that many cellulase molecules.
- When produced, a cellulase molecule either:
- stays where it was produced [LEFT SIMULATION] or
- floats a short distance randomly [RIGHT SIMULATION]
- Each iteration, some random bacteria are given a chance to reproduce.
- Cells with higher food supply have a greater chance to reproduce.
- Food supply is defined as the amount of cellulase present at the cell.
- Reproduction means:
- Overwriting a nearby cell with one's own genotype.
- Reproduction is not exact. There is "mutation".
- The offspring's genotype is a number close to the parent's.
The left half of the simulation models the case where the cellulase produced by a cell can only benefit itself. The right half models the case where the cellulase can float away and benefit neighbours instead of the cell that produced it.
C code
C code for the simulator is available. Note that, by default, it will spit out about 62 MB of .bmp graphics files so you can see visually what is going on.
A much faster, but less mathematically rigorous version, is also available.
Simulation run
In this simulation, each pixel represents a bacterium. Its brightness indicates its ability to produce cellulase. In the left half of the simulation, the amount of cellulase available to a cell is entirely dependent on its genotype, whereas in the right half, the amount also depends on the genotype of nearby cells.
Graph
Here's a graph of the simulation...
Conclusion
This is an excellent reason why surface display of cellulases is preferable to releasing them into the media. Surface display drastically improves the prospects for simply evolving the desired phenotype.
References
- Van Zyl WH, Lynd LR, Den Haan R, McBride EJ (2007) [http://www.springerlink.com/content/4l4m28lp06120253/ Consolidated bioprocessing for bioethanol production using Saccharomyces cerevisiae]. Advances in Biochemical Engineering/Biotechnology 108: 205-235 (doi: 10.1007/10_2007_061).
 "
"