|
|

E. coli
Construction of RSR+Leader Array
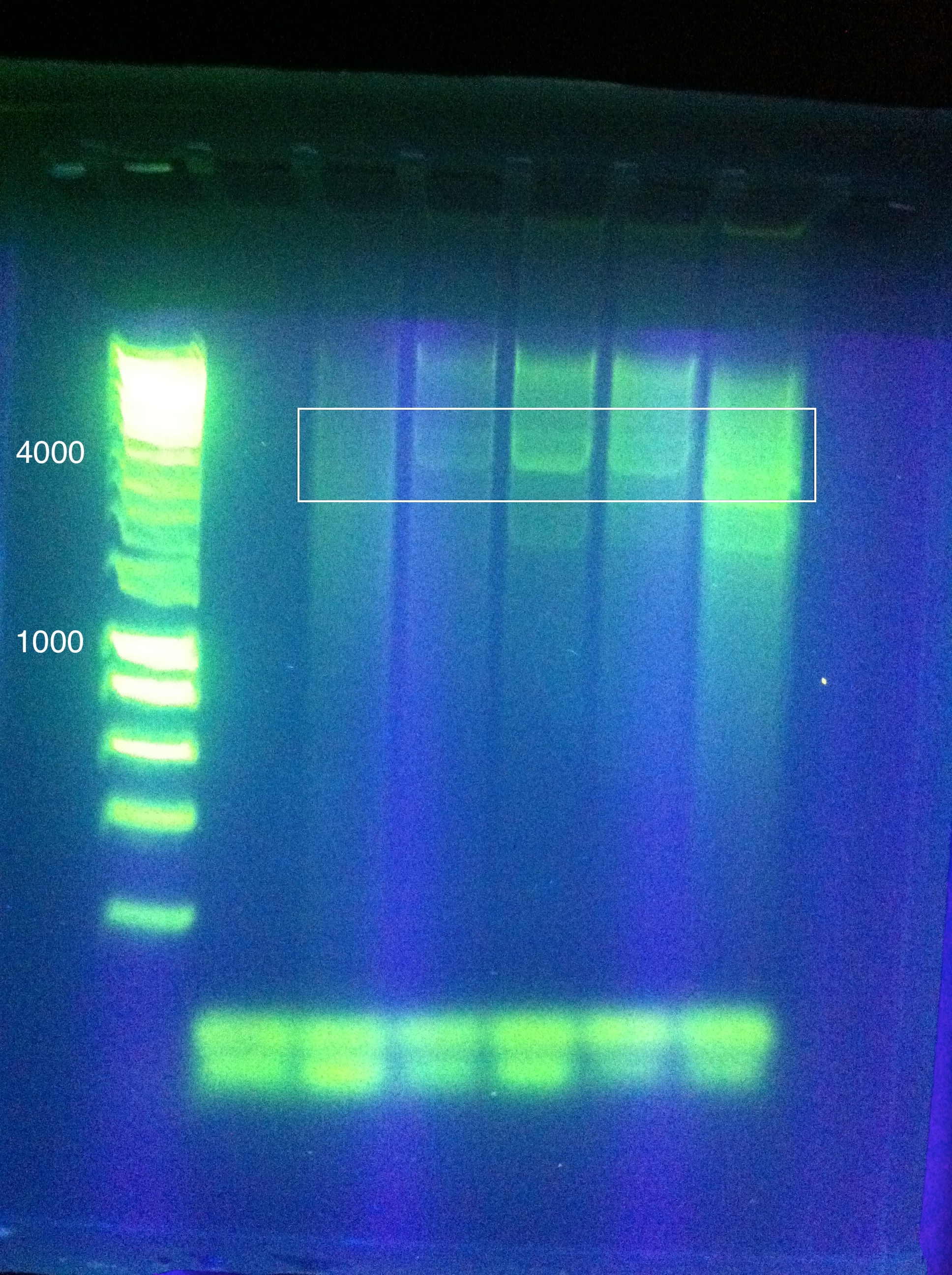
- Gel results: Correct size for the RSR+Leader Array in the 6th well. Confirmed with [http://sequencing.biodesign.asu.edu/ DNASU] sequencing.
- Sequencing verification showed a 99% base pair match for "Seq1" and the MG1655 leader sequence, confirming the successful assembly of the array
Isolation of Cas genes
- CasABCDE was unsuccessful, though we came close:
- Cas3 was moderately successful, but unconfirmed via sequencing:
Assembly of Leader+RSR into pRSF Duet
-
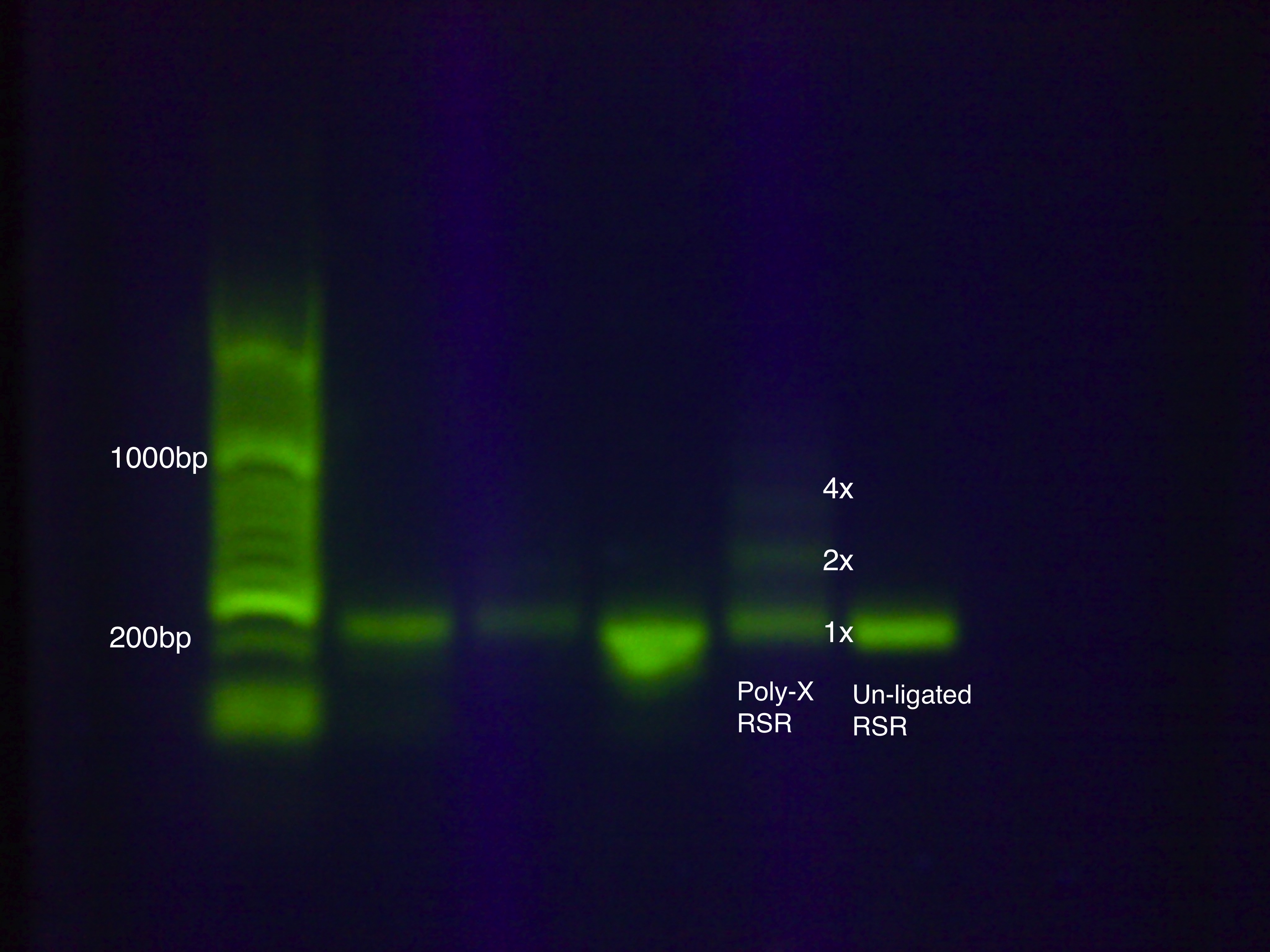
We devised a new assembly method, the Poly-X Ligation, for RSR construction and tested it on an RSR construct with GFP.
Contingency Test
- Standardized competent cells
- Plate results:
Absorbance Conditions
| 96 Well Plate Type
| Corning Costar
|
| Well Surface Area
| 0.32 cm2
|
| Sample Volume
| 0.1 cm3
|
| Path Distance
| 0.3125 cm
|
Pre-Competency Preparation
| Culture
| Raw Absorbance / Recorded OD600*
| Mean OD600
|
| BL21 L+Seq1 | 0.216 | 0.302
0.965 | 0.309
0.987 | | 0.976
|
| 1655 L+Seq1 | 0.343
1.098 | 0.351
1.122 | 0.354
1.131 | | 1.117
|
| BL21 Leader | 0.259
0.830 | 0.271
0.867 | 0.272
0.871 | | 0.856
|
| 1655 Leader | 0.224
0.716 | 0.215
0.688 | 0.215
0.689 | | 0.697
|
| BL21 GFP | 0.317
1.014 | 0.342
1.096 | 0.321
1.026 | | 1.045
|
| 1655 GFP | 0.127
0.406 | 0.137
0.438 | 0.125
0.400 | | 0.415
|
| trash | 0.046
0.148 | 0.047
0.149 | 0.046
0.148 | | 0.148
|
| trash | 0.046
0.148 | 0.047
0.149 | 0.047
0.149 | | 0.148
|
| Amp blank | 0.039
0.123 | 0.041
0.131 | 0.040
0.127 | 0.040
0.128 | 0.127
|
| Kan blank | 0.040
0.127 | 0.040
0.128 | 0.041
0.131 | 0.040
0.128 | 0.129
|
*Recorded OD600 = raw absorbance * 3.2
Competency Prep Dilution
(1/100th dilution, grown for 2 hours)
| Culture
| Raw Absorbances / Recorded OD600
| Mean OD600
|
| BL21 L+Seq1 | 0.081
0.260 | 0.079
0.252 | 0.083
0.265 | 0.259
|
| 1655 L+Seq1 | 0.077
0.245 | 0.078
0.250 | 0.085
0.272 | 0.256
|
| BL21 Leader | 0.068
0.218 | 0.072
0.229 | 0.077
0.246 | 0.231
|
| 1655 Leader | 0.060
0.193 | 0.061
0.195 | 0.062
0.198 | 0.196
|
| BL21 GFP | 0.054
0.171 | 0.054
0.172 | 0.055
0.175 | 0.173
|
| 1655 GFP | 0.047
0.149 | 0.048
0.153 | 0.048
0.152 | 0.151
|
Transformation Efficiency Results
| E. Coli Strain
| Initial Plasmid
| Introduced Plasmid
| Antibiotic selection
| DNA Quantity Used (ng)
| # of Colonies
| Weighted Colony Count**
|
| MG1655 | GFP Construct in pRSF | Leader in pIDT | Amp | 140 | 21 | 138.742
|
| MG1655 | GFP Construct in pRSF | Leader+Seq1 in pIDT | Amp | 147 | 31 | 204.810
|
| MG1655 | Leader in pIDT | GFP Construct in pRSF | Kan | 143.4 | 23 | 117.635
|
| MG1655 | Leader+Seq1 in pIDT | GFP Construct in pRSF | Kan | 143.4 | 1000 | 3909.508
|
| BL21 | GFP Construct in pRSF | Leader in pIDT | Amp | 140 | 0 | 0.000
|
| BL21 | GFP Construct in pRSF | Leader+Seq1 in pIDT | Amp | 147 | 2 | 11.574
|
| BL21 | Leader in pIDT | GFP Construct in pRSF | Kan | 143.4 | 14 | 60.512
|
| BL21 | Leader+Seq1 in pIDT | GFP Construct in pRSF | Kan | 143.4 | 9 | 35.186
|
**Weighted Colony Count = (# of Colonies)/(mean OD600 of competency prep)
B. halodurans
Construction of RSR Array
- We constructed a 1x Repeat-Spacer-Repeat array by ligating our "RA" (Spacer-Repeat) sequence to our "RB" (Repeat) sequence
Isolation of cmr genes
- Our construct requires the isolation of 6 genes, Cmr1-6, that are all located on a single locus.
- Gel results (CMR success):
Assembly into pRSF Duet
L. innocua
Construction of RSR Array
- This construction of the RSR array involved the ligation of two customized BioBricks (sequences), which can be done simultaneously using this protocol.
Cas gene
- The L. innocua CRISPR system utilizes three CRISPR-associated genes: Cas1, Cas2, and Cas9.
- For our streamlined construct, we needed to isolate only Cas9, which is approximately 4kb in length.
- We PCR amplified Cas9 with the adjacent trans-encoded CRIPSR RNA region (tracrRNA), which is necessary for the formation of mature CRISPR RNA.
Assembly into pRSF Duet
- Cas9+tracrRNA array has been ligated into pRSF Duet
Other
- Construction of constitutive GFP plasmid by ligation of Bba_E0840 with Bba_(promoter)
- Success was marked by green colonies
|
 "
"