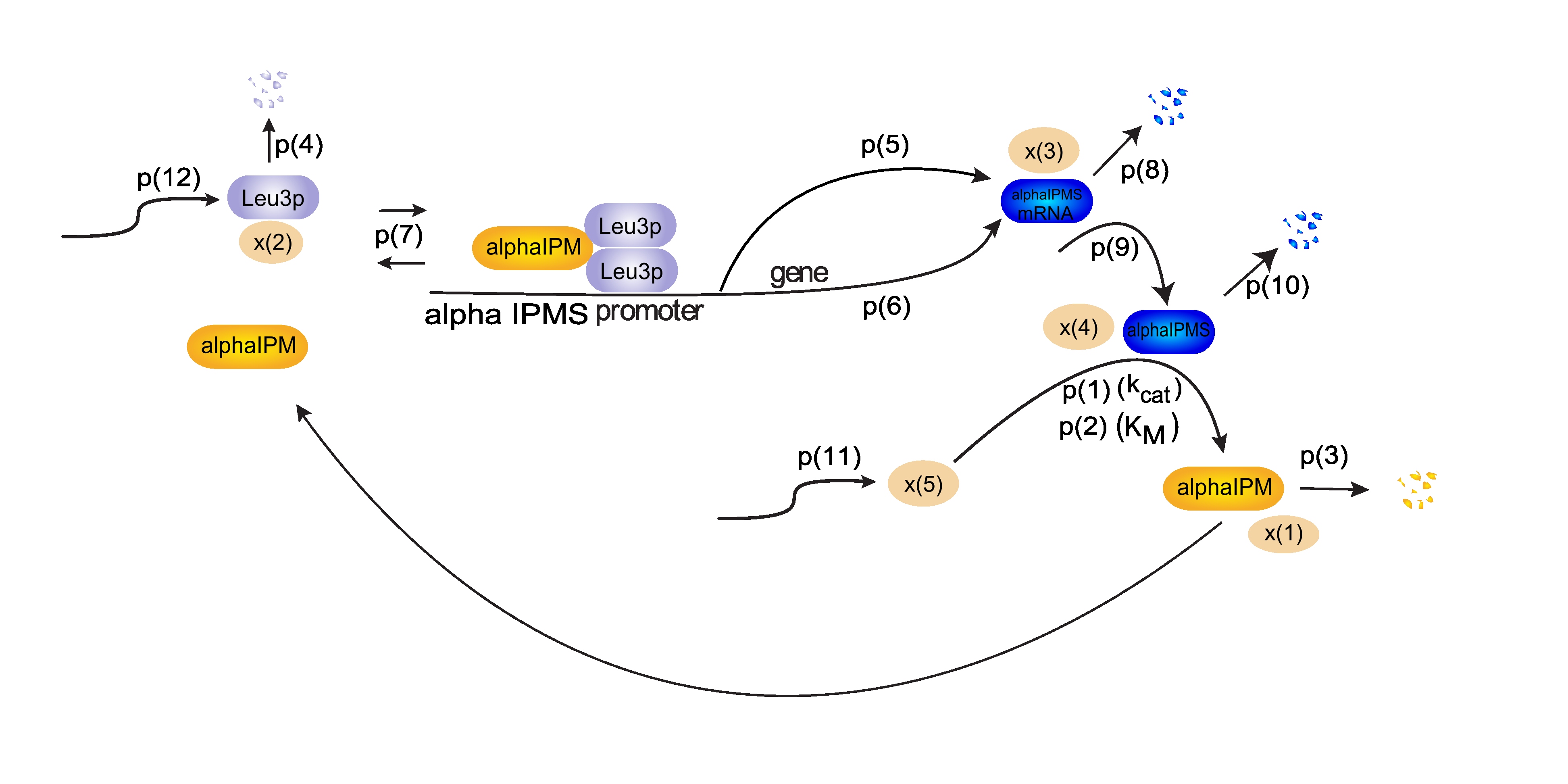
The Leu3p (x(2)) dimer blocks transcription from the alpha-IPM synthase (alpha IPMS) promoter. Alpha-IPM (x(1)) binding to this dimer releases it from the promoter and enhances the transcription rate. p(5) and p(6) model both constitutive expression and the transcription rate increase upon alpha-IPM binding. From alpha-IPMS mRNA (x(3)) the protein is formed, modelled by p(9). Alpha-IPMS (x(4)) enzymatically produces alpha-IPM, which is then incorporated in the beginning of this loop again. Intercellular transport of alpha-IPM creates a signal transduction through a hypha.
Additionally, x(5) is required for modelling the enzymatic reaction. Its formation is described by p(11). Each type of molecule has also been assigned a degradation constant, such as p(8) for x(3).
 "
"