Parts
Our Biobrick 1: BBa_K512000
 |
|
|
|
|
E. curi
|
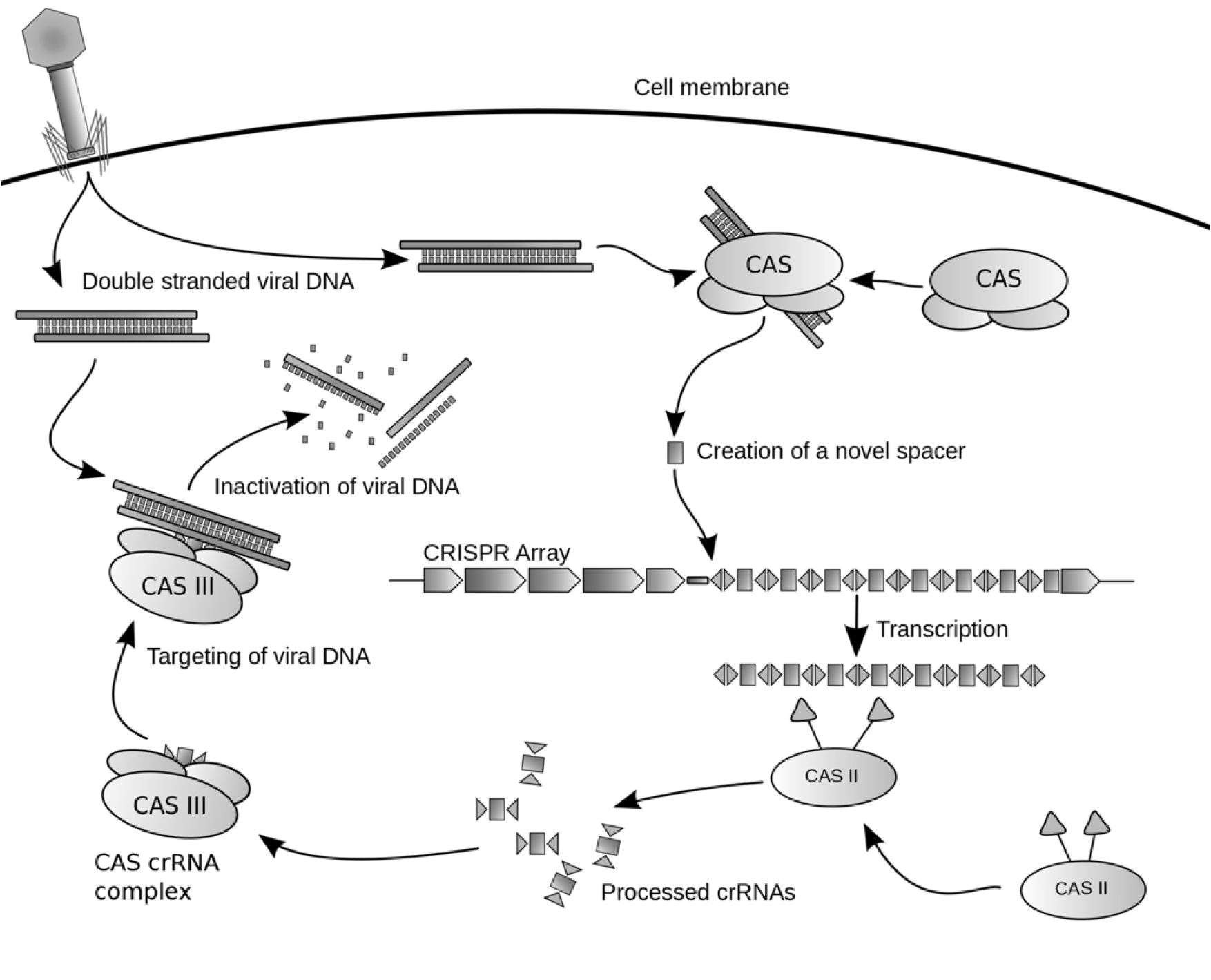
Bacteria protect their genome and remove foreign DNA elements through the use of the clustered, regularly interspaced short palindromic repeat (CRISPR) system. Recently discovered, this defense mechanism can protect microorganisms against bacteriaphages. The CRISPR system comprises a transcribed locus of repeats and spacers that is activated and made functional via a family of CAS proteins. The CRISPR repeats are short DNA sequences that are almost identical in size and sequence. Between a pair of repeats is a spacer, which if complementary to a DNA element of the invading bacteriaphage, facilitates the destruction of that sequence from the bacterial cell. As such, the CRISPR/CAS system is a primitive biological defense mechanism, similar in many ways to the process of adaptive immunity in eukaryotic immune systems. Plasmids, which are commonly used in molecular biology are also the source of horizontal gene transfer in the wild and facilitate the spread of antibiotic resistant genes among microorganisms. These plasmids can contain unique DNA sequences that could be targeted by the CRISPR/CAS system. We intend to exploit the CRISPR/CAS system to engineer a mechanism of plasmid curing and deactivation of antibiotic resistance genes in E. coli.
|
|
|
|
We synthesized a version of CRISPR encoding a spacer that matches the GFP DNA sequence. We tested the synthetic CRISPR array against E.coli harboring a tetO::GFP plasmid that confers ampicillin resistance. Activation of CRISPER-GFP destroys the GFP-containing plasmid restoring the bacterial host's sensitivity to ampicillin. We will use the synthetic CRISPR system as a biological tool, combining it with other BioBricks for use in applications that will impact health and medicine, biotechnology, molecular biology, and genetics.
|
 | Curran Laboratory at the USC School of Gerontology
|
| File:Labofmicriobio wageningen.gif | Brouns Laboratory at the University of Wageningen
|
 | Integrated DNA Technologies
|
 | David and Dana Dornsife College of Arts and Sciences
|
| File:Viterbi.gif | Viterbi School of Engineering
|
|
|
|
We've focussed on building one key new BioBrick; BBa_K512000; encoding cas3 genes which can improve CRIPSR. During the project we both made and well characterised this part, details of which can be found in our Notebook section, whilst information on its sequence and how to get hold of it can be found at the [http://partsregistry.org/Part:BBa_K512000] page on the parts registry website.
Our Biobrick 2: BBa_K512001
 |
|
|
|
|
E. curi
|
Bacteria protect their genome and remove foreign DNA elements through the use of the clustered, regularly interspaced short palindromic repeat (CRISPR) system. Recently discovered, this defense mechanism can protect microorganisms against bacteriaphages. The CRISPR system comprises a transcribed locus of repeats and spacers that is activated and made functional via a family of CAS proteins. The CRISPR repeats are short DNA sequences that are almost identical in size and sequence. Between a pair of repeats is a spacer, which if complementary to a DNA element of the invading bacteriaphage, facilitates the destruction of that sequence from the bacterial cell. As such, the CRISPR/CAS system is a primitive biological defense mechanism, similar in many ways to the process of adaptive immunity in eukaryotic immune systems. Plasmids, which are commonly used in molecular biology are also the source of horizontal gene transfer in the wild and facilitate the spread of antibiotic resistant genes among microorganisms. These plasmids can contain unique DNA sequences that could be targeted by the CRISPR/CAS system. We intend to exploit the CRISPR/CAS system to engineer a mechanism of plasmid curing and deactivation of antibiotic resistance genes in E. coli.
|
|
|
|
We synthesized a version of CRISPR encoding a spacer that matches the GFP DNA sequence. We tested the synthetic CRISPR array against E.coli harboring a tetO::GFP plasmid that confers ampicillin resistance. Activation of CRISPER-GFP destroys the GFP-containing plasmid restoring the bacterial host's sensitivity to ampicillin. We will use the synthetic CRISPR system as a biological tool, combining it with other BioBricks for use in applications that will impact health and medicine, biotechnology, molecular biology, and genetics.
|
 | Curran Laboratory at the USC School of Gerontology
|
| File:Labofmicriobio wageningen.gif | Brouns Laboratory at the University of Wageningen
|
 | Integrated DNA Technologies
|
 | David and Dana Dornsife College of Arts and Sciences
|
| File:Viterbi.gif | Viterbi School of Engineering
|
|
|
|
We also builded another key new BioBrick; BBa_K512001; encoding casABCDE12 genes which is a series of cas genes responsible for CRISPR interference to work in E. coli. During the project we both made and well characterised this part, details of which can be found in our Notebook section, whilst information on its sequence and how to get hold of it can be found at the [http://partsregistry.org/Part:BBa_K512001] page on the parts registry website.
Our Biobrick 2: BBa_K512002
 |
|
|
|
|
E. curi
|
Bacteria protect their genome and remove foreign DNA elements through the use of the clustered, regularly interspaced short palindromic repeat (CRISPR) system. Recently discovered, this defense mechanism can protect microorganisms against bacteriaphages. The CRISPR system comprises a transcribed locus of repeats and spacers that is activated and made functional via a family of CAS proteins. The CRISPR repeats are short DNA sequences that are almost identical in size and sequence. Between a pair of repeats is a spacer, which if complementary to a DNA element of the invading bacteriaphage, facilitates the destruction of that sequence from the bacterial cell. As such, the CRISPR/CAS system is a primitive biological defense mechanism, similar in many ways to the process of adaptive immunity in eukaryotic immune systems. Plasmids, which are commonly used in molecular biology are also the source of horizontal gene transfer in the wild and facilitate the spread of antibiotic resistant genes among microorganisms. These plasmids can contain unique DNA sequences that could be targeted by the CRISPR/CAS system. We intend to exploit the CRISPR/CAS system to engineer a mechanism of plasmid curing and deactivation of antibiotic resistance genes in E. coli.
|
|
|
|
We synthesized a version of CRISPR encoding a spacer that matches the GFP DNA sequence. We tested the synthetic CRISPR array against E.coli harboring a tetO::GFP plasmid that confers ampicillin resistance. Activation of CRISPER-GFP destroys the GFP-containing plasmid restoring the bacterial host's sensitivity to ampicillin. We will use the synthetic CRISPR system as a biological tool, combining it with other BioBricks for use in applications that will impact health and medicine, biotechnology, molecular biology, and genetics.
|
 | Curran Laboratory at the USC School of Gerontology
|
| File:Labofmicriobio wageningen.gif | Brouns Laboratory at the University of Wageningen
|
 | Integrated DNA Technologies
|
 | David and Dana Dornsife College of Arts and Sciences
|
| File:Viterbi.gif | Viterbi School of Engineering
|
|
|
|
This is our most important BioBrick; BBa_K512002; which is the CRISPR system with GFP target spacer we use during our research. It can target GFP plasmid in E. Coli cells and degrade it. Again, during the project we both made and well characterised this part, details of which can be found in our Notebook section, whilst information on its sequence and how to get hold of it can be found at the [http://partsregistry.org/Part:BBa_K512002] page on the parts registry website.
|

 "
"