Results
Contents |
Parts assembly
All the parts have been cloned with success. The part name, plasmids and quality controls are reported in the Freezer section.Characterization of basic modules
Characterization of promoters pTet and pLux
Inducible and constitutive promoters were assembled upstream of different coding sequences containing an RBS from the Community collection.
The assembled RBSs are:
| BioBrick code | Declared efficiency |
| BBa_B0030 | 0,6 |
| BBa_B0031 | 0,07 |
| BBa_B0032 | 0,3 |
| BBa_B0034 | 1 |
For an inducible device, the RBS variation has the purpose to stretch the induction curve, thus modulating its PoPs-OUT range.
The complex RBS-promoter acts as a whole regulatory element and determines the amount of translated protein. RBSs have been reported to have an un-modular behavior, since the translational efficiency is not independent on the coding sequences, but variates as an effect of different mRNA structure stability [Salis et al., Nat Biotec, 2009]. It is not possible to separate the effects of the sole promoter and of the sole RBS on the total amount/activity of gene product (in this case study, mRFP).
For this reason, every combination 'Promoter+RBS' was studied as a different regulatory element. Regulatory elements were characterized using mRFP reporter protein for different RBSs in terms of Synthesis rate per Cell (Scell) and R.P.U.s (Relative Promoter Units) as explained in measurements section.
Operative parameters of the promoter are derived from the estimated Hill equations obtained by nonlinear least squares fitting (lsqnonlin Matlab routine) of the Hill function expressed in RPUs:
- RPUmax is equal to the α and represents the maximum promoter activity
- RPUmin is equal to the α * δ represents the minimum promoter activity
- Switch point is computed as the abscissa of the inflection point of the Hill curve and it is representative of the position of linear region
- Linearity boundaries are determined as the intersection between the tangent line to the inflection point and the upper and lower horizontal boundaries of the Hill curve.
The estimated parameters for the Hill functions of pLux are summarized in the table below. For more details on parameter estimation, see the model section.
| RBS | αpLux [(AUr/min)/cell] | δpLux [-] | ηpLux [-] | kpLux [ng/ml] |
| BBa_B0030 | 438 [10] | 0.05 [>100] | 2 [47] | 1.88 [27] |
| BBa_B0031 | 9.8 [7] | 0.11 [57] | 1.2 [29] | 1.5 [26] |
| BBa_B0032 | 206 [3] | 0 [>>100] | 1.36 [10] | 1.87 [9] |
| BBa_B0034 | 1105 [6] | 0.02 [>100] | 1.33 [19] | 2.34 [18] |
The operative parameters are summarized in the table below:
| RBS | RPUmax | RPUmin | Switch point [nM] | Linear boundaries [MIN; MAX] [nM] |
| B0030 | 4.28 | 0.20 | 1.08 | [0.36; 3.27] |
| B0031 | 4.93 | 0.55 | 0.25 | [0.03; 2.30] |
| B0032 | 9.49 | 0.02 | 0.47 | [0.07; 3.07] |
| B0034 | 21.53 | 0.51 | 0.53 | [0.08; 3.77] |
The protocols for the characterization of pTet promoter are reported in the pTet measurement section.
Here we have characterized its transcriptional strength as a function of aTc induction (ng/ul) for different RBSs. Three different induction curves were obtained and are reported in figure:
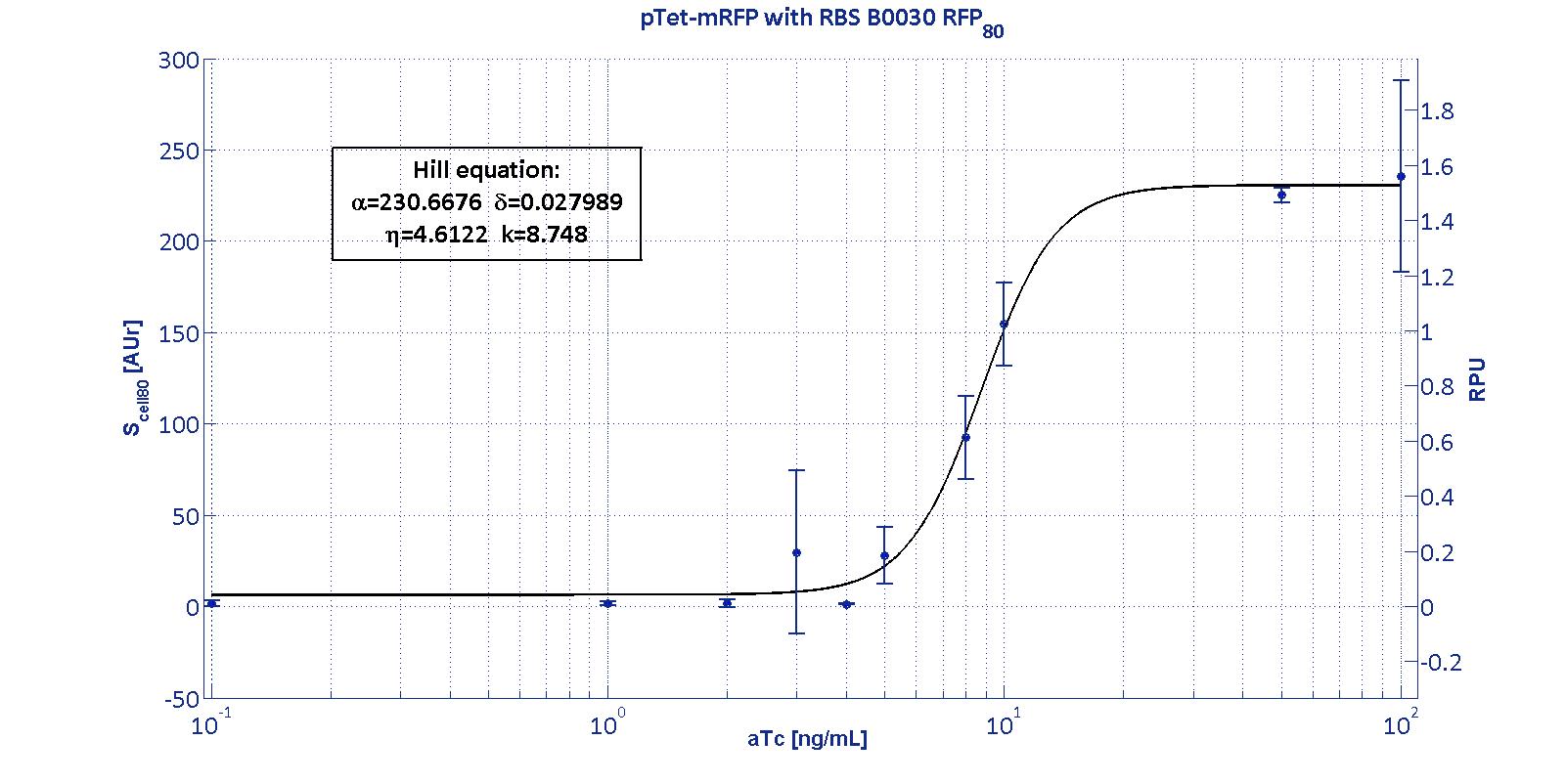
The estimated parameters of the Hill curves described in the figures are summarized in the table below:
| RBS | αpTet [(AUr/min)/cell] | δpTet [-] | ηpTet [-] | kpTet [nM] |
| BBa_B0030 | 230.67 [3.7] | 0.028 [91.61] | 4.61 [23.73] | 8.75 [4.16] |
| BBa_B0031 | ND | ND | ND | ND |
| BBa_B0032 | 55.77 [12] | 1.53E-11 [>>100] | 4.98 [57.62] | 7.26 [14.98] |
| BBa_B0034 | 120 [5.95] | 0.085 [40.6] | 24.85 [47.6] | 9 [5.43] |
The operative parameters are summarized in the table below:
| RBS | RPUmax | RPUmin | Switch point [ng/ml] | Linear boundaries [MIN; MAX] [ng/ml] |
| B0030 | 1.53 | ~0 | 7.95 | [4.66;11.99] |
| B0031 | ND | ND | ND | ND |
| B0032 | 3.16 | ~0 | 6.7 | [4.45;10.05] |
| B0034 | 2.73 | 0.23 | 8.96 | [8.27;9.71] |
 "
"





