Characterized Parts
NB: unless differently specified, all tests were performed in E. coli MGZ1 in M9 supplemented medium at 37°C in low copy plasmid pSB4C5.
New parts
J23101x series
- BBa_K516130 (wiki name: J101-E5 ) J101-RBS30-mRFP-TT
- BBa_K516131 (wiki name: J101-31 ) J101-RBS31-mRFP-TT
- BBa_K516132 (wiki name: J101-E7 ) J101-RBS32-mRFP-TT
BBa_J23101 is the reference standard promoter for the computation of RPUs. As discussed in 'data analysis' section, RPUs are relative units for the evaluation of promoter strength, based on a mathematical model of the transcription and the translation of a reporter gene.
The RPUs are supposed to be indepedent on the experimental setup, provided that the reference standard BBa_J23101 must be assayed in the same experimental condition of the studied promoter. It means that if the studied promoter is in a low copy number plasmid and drives the expression of a reporter protein P, J23101 must be assembled in the same vector upstream of the same reporter P. This approach is in accordance with the philosophy of synthetic biology, based on the concept of 'modularity' of the components. According to this approach, the assembly of basic well characterized modules to build complex circuits allows the prediction of the circuit behavior starting from the knowledge on the basic parts.
Salis et al. [Nat Biotec, 2009] stated that 'Identical ribosome binding site sequences in different genetic contexts can result in different protein expression levels' and again 'It is likely that this absence of modularity is caused by the formation of strong secondary structures between the RBS-containing RNA sequence and one protein coding sequence but not another.'
For this reason, RPUs might not be reliable when comparing the same promoter with different RBSs because of the un-modularity of the RBS. In order to asses what's the effect of RBS 'un-modularity' on RPUs reliability, we have built a set of four constitutive promoters (BBa_J23101) followed by one of the four RBSs tested. These parts were used to evaluate RBS efficiency. Data were collected and analyzed as described in 'Measurements' and 'Data analysis' sections. RPUs and Synthesis rate per cell [AUr] were computed and results are summarized in the table below.
NB: in the RPU computation, the J23101-RBS34-mRFP-TT (BBa_J23101) in low copy plasmid pSB4C5 construct has been considered as the reference standard. With this assumption, RPUs are identical to the estimated RBS efficiency.
|
LuxI expression cassette driven by aTc-inducible pTet promoter
- BBa_K516210 (wiki name: E13 ) pTet-RBS30-LuxI
- BBa_K516211 (wiki name: E14 ) pTet-RBS31-LuxI
- BBa_K516212 (wiki name: E15 ) pTet-RBS32-LuxI
- BBa_K516214 (wiki name: E16 ) pTet-RBS34-LuxI
Though these parts don't have a transcriptional terminator, they have been characterized in low copy plasmid pSB4C5, that contains the BBa_B0054 terminator. This choice is motivated by the need to reproduce the exact experimental context of the final circuit, as described in solution section.
LuxI has been characterized in terms of enzymatic activity under the regulation of pTet promoter.
KM,LuxI and Vmax parameters representing its activity have been estimated and the promoter strength (represented by a synthetic parameter αpTet for every pTet-RBS combination) at full induction (100 ng/ml) has been estimated too with a simultaneous fitting of the available data.
LuxI has been characterized through the Biosensor BBa_T9002 (see modelling section).
The HSL synthesis rate has been evaluated according to the model equations, properly adjusted. In particular, the ODE system describing the behavior of this measurement circuit is reported here:
Since the measurement systems are only assayed in the exponential growth phase, in the equation (3) N < Nmax is assumed. The parameters Nmax, μ and γHSL (see the Results section for more details) are known and summarized in the table below:
| Nmax | μ | γHSL |
| 1*109 | 0.004925 | 0 |
The parameters Vmax, kM,LuxI and αRBSx were estimated with a simultaneous fitting of the data collected as described in measurement section for the four measurement parts pTet-RBSx-LuxI-TT assayed by BBa_T9002 biosensor section.
The estimated parameters for the enzymatic activity of LuxI are reported in the table below:
| Vmax | kM,LuxI | αB0030 | αB0031 | αB0032 | αB0034 |
| 3.56*10-9 | 6.87*103 | 87 | 8.5 | ND | 252 |
The collected data have been used to identify the parameters of our model. Despite the data-poor context, the model predictions fit the experimental data, thus demonstrating that the equation that models the HSL synthesis by LuxI is a good approximation of real processes.
AiiA expression cassette driven by aTc-inducible pTet promoter
- BBa_K516220 (wiki name: E24 ) pTet-RBS30-AiiA-TT
- BBa_K516221 (wiki name: E25 ) pTet-RBS31-AiiA-TT
- BBa_K516222 (wiki name: E26 ) pTet-RBS32-AiiA-TT
- BBa_K516224 (wiki name: E27 ) pTet-RBS34-AiiA-TT
The AiiA enzyme activity has been characterized under the regulation of ptet promoter, assaying its enzymatic activity. Similar to LuxI, a system of differential equations (referring to model equations) has been derived.
The parameters kcat, kM,AiiA and αRBSx would have been estimated with a simultaneous fitting of the data collected as described in measurement section for the four measurement parts pTet-RBSx-AiiA-TT assayed by BBa_T9002 biosensor section.
Unfortunately, their estimation revealed difficult.
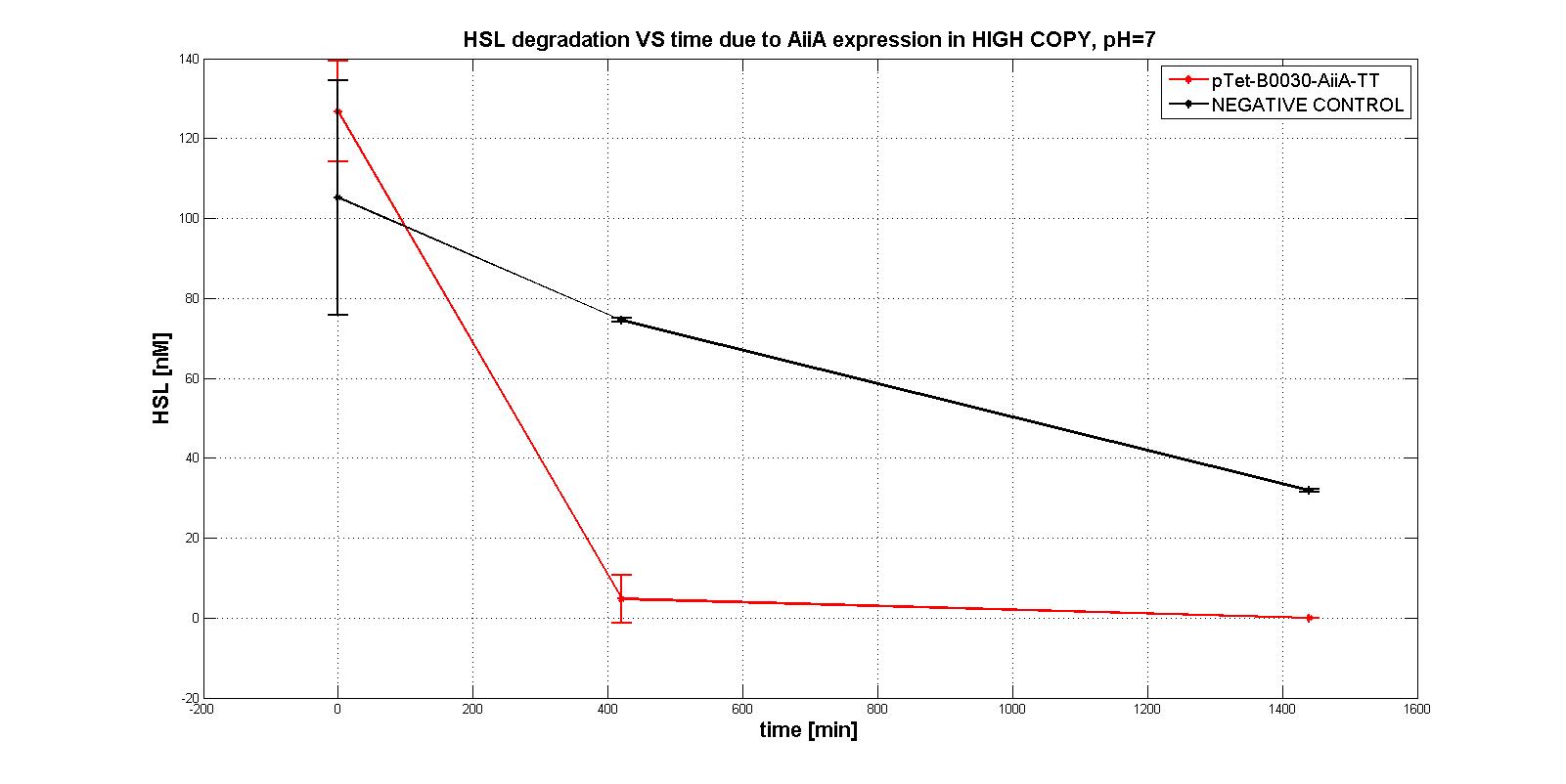
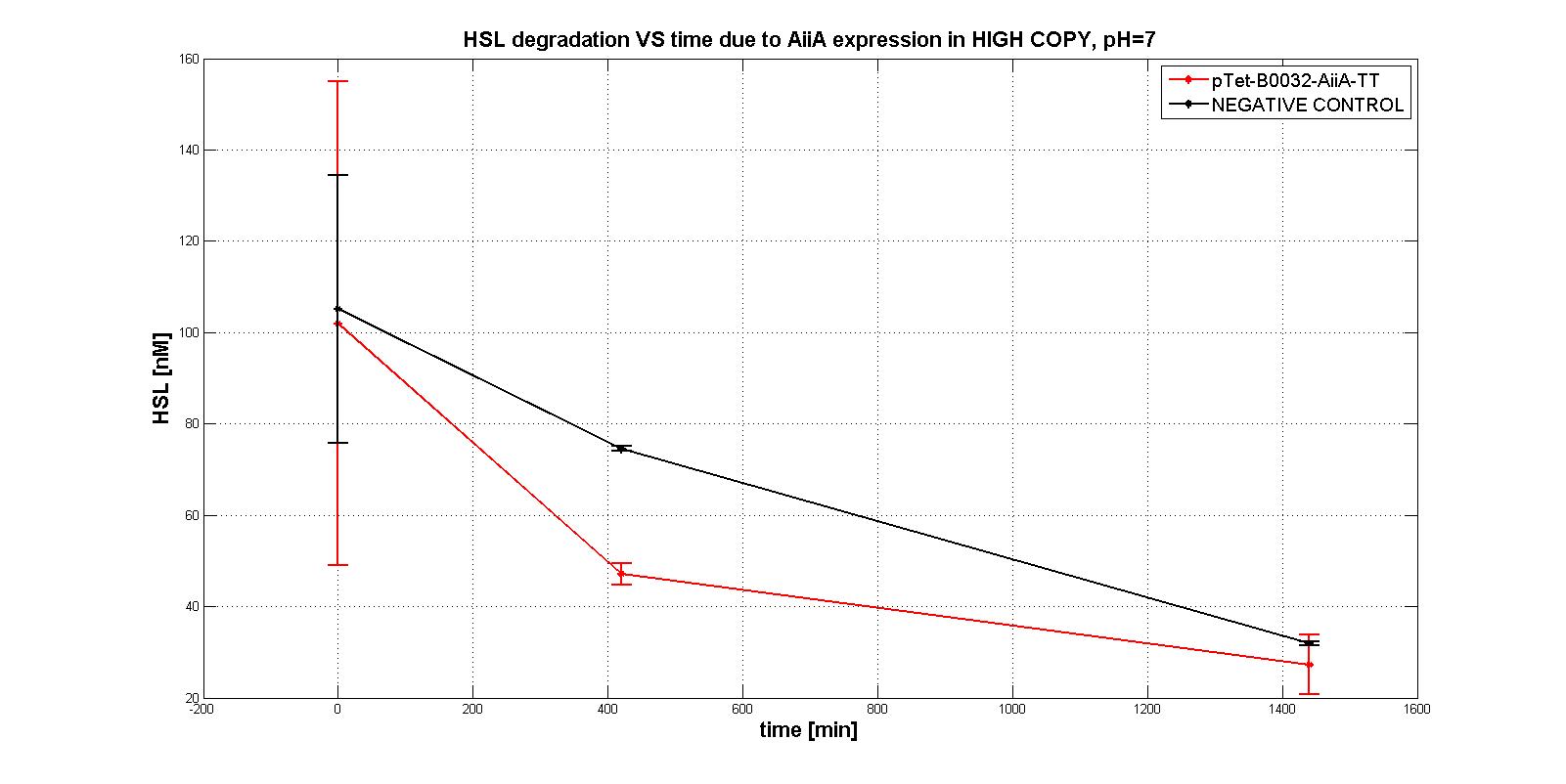
In the first experiments with the measurement system pTet-RBSx-AiiA-TT in LOW-COPY at pH=7 no degradation of HSL was observed. The collected data are shown in the figure below. HSL degradation is identical in the measurement system and in the negative control after 21 hours.
Existing parts
Notes for promoter characterization
Inducible and constitutive promoters were assembled upstream of different coding sequences containing an RBS from the Community collection.
The assembled RBSs are:
| BioBrick code | Declared efficiency |
| BBa_B0030 | 0,6 |
| BBa_B0031 | 0,07 |
| BBa_B0032 | 0,3 |
| BBa_B0034 | 1 |
For an inducible device, the RBS variation has the purpose to stretch the induction curve, thus modulating its PoPs-OUT range.
The complex RBS-promoter acts as a whole regulatory element and determines the amount of translated protein. RBSs have been reported to have an un-modular behavior, since the translational efficiency is not independent on the coding sequences, but variates as an effect of different mRNA structure stability [Salis et al., Nat Biotec, 2009]. It is not possible to separate the effects of the sole promoter and of the sole RBS on the total amount/activity of gene product (in this case study, mRFP).
For this reason, every combination 'Promoter+RBS' was studied as a different regulatory element. Regulatory elements were characterized using mRFP reporter protein for different RBSs in terms of Synthesis rate per Cell (Scell) and R.P.U.s (Relative Promoter Units) as explained in measurements section.
Operative parameters of the promoter are derived from the estimated Hill equations obtained by nonlinear least squares fitting (lsqnonlin Matlab routine) of the Hill function expressed in RPUs:
- RPUmax is equal to the α and represents the maximum promoter activity
- RPUmin is equal to the α * δ represents the minimum promoter activity
- Switch point is computed as the abscissa of the inflection point of the Hill curve and it is representative of the position of linear region
- Linearity boundaries are determined as the intersection between the tangent line to the inflection point and the upper and lower horizontal boundaries of the Hill curve.
RBSs
The evaluation of RBS efficiency can be performed in a very intuitive fashion:
- select the RBSs you want to study
- assemble them in a Promoter - XX - Coding sequence circuit
- measure the output of the circuits and calculate the RBS efficiency as the ratio of the output relative to the output of the circuit with the standard RBS (BBa_B0034).
 |
This simple measurement system allows the quantification of RBS efficiency depending on the experimental context (i.e.: promoter and encoded gene). Today it has not still been completely validated the hypothesis that every functional module in a genetic circuit maintains its behavior when assembled in complex circuits, even if many researchers implicitly accept this hypothesis when performing characterization experiments.
To rationally assess the impact that this hypothesis has on the genetic circuit design and fine tuning, several measurement systems were built to evaluate the dependance of RBS modularity from the promoter or the coding sequence separately.
In particular, in order to investigate if RBS efficiency depends on the promoter, the same coding devices (RBSx-RFP-TT) were assembled downstream of different promoters (J23101, pTet, pLux). The system output was measured and the RBS efficiency evaluated. The results are summarized in the table below:
| RBS | effpLux* | effpTet* | effJ23101** | Declared efficiency |
| B0030 | 0.40 | 1.6814 | 2.45 | 0,6 |
| B0031 | 0.01 | ND | 0.04 | 0,07 |
| B0032 | 0.19 | 0.4193 | 0.40 | 0,3 |
| B0034 | 1 | 1 | 1 | 1 |
| RBS | effmRFP** | effAiiA*** | effLuxI**** | Declared efficiency |
| B0030 | 1.72 | 0.53 | 0.45 | 0,6 |
| B0031 | 0.03 | 0.83 | 0.028 | 0,07 |
| B0032 | 0.37 | 0.50 | N.D. | 0.3 |
| B0034 | 1 | 1 | 1 | 1 |
* The RBS efficiency for inducible devices expressing mRFP was estimated as the ratio of the AUCs (Area under the curve) of the induction curve of the system with the studied RBS and the B0034 reference: AUCP, RBSx/AUCP, B0034
** The RBS efficiency for constitutive promoters expressing mRFP was computed as the ratio between ScellP, RBSx/ScellP, B0034
*** The RBS efficiency for pTet promoter driving the expression of AiiA enzyme was computed as the ratio between the percentage of degraded HSL after 4 hours of the studied system and the reference B0034. pTet was tested at full induction (100 ng/ml).
**** The RBS efficiency for promoters driving the expression of LuxI was computed as the ratio αpTet, RBSx/αpTet, B0034 estimated from the measurement systems pTet-RBSx-LuxI. αpTet was estimated as described here. pTet was tested at full induction (100 ng/ml).
The parts we used to characterize the RBSs are listed here:
- mRFP expression with different promoters
- J23101
- BBa_K516130 J101-RBS30-RFP-TT
- BBa_K516131 J101-RBS31-mRFP-TT
- BBa_K516132 J101-RBS32-mRFP-TT
- BBa_J23101 J101-RBS34-mRFP-TT
- pTet
- BBa_K516230 pTet-RBS30-mRFP-TT
- BBa_K516231 pTet-RBS31-mRFP-TT
- BBa_K516232 pTet-RBS32-mRFP-TT
- BBa_I13521 pTet-RBS34-mRFP-TT
- pLux
- BBa_K516330 pLambda-RBS30-LuxR-T-pLux-RBS30-mRFP-TT
- BBa_K516331 pLambda-RBS30-LuxR-T-pLux-RBS31-mRFP-TT
- BBa_K516332 pLambda-RBS30-LuxR-T-pLux-RBS32-mRFP-TT
- BBa_K516334 pLambda-RBS30-LuxR-T-pLux-RBS34-mRFP-TT
- pTet driving the expression of different genes
- mRFP
- BBa_K516230 pTet-RBS30-mRFP-TT
- BBa_K516231 pTet-RBS31-mRFP-TT
- BBa_K516232 pTet-RBS32-mRFP-TT
- BBa_I13521 pTet-RBS34-mRFP-TT
- AiiA
- BBa_K516220 pTet-RBS30-AiiA-TT
- BBa_K516221 pTet-RBS31-AiiA-TT
- BBa_K516222 pTet-RBS32-AiiA-TT
- BBa_K516224 pTet-RBS34-AiiA-TT
- LuxI
- BBa_K516210 pTet-RBS30-LuxI
- BBa_K516211 pTet-RBS31-LuxI
- BBa_K516212 pTet-RBS32-LuxI
- BBa_K516214 pTet-RBS34-LuxI
The results reported in the table suggest that the RBS efficiency ranking is not always maintained. In particular, for the different promoters driving the expression of mRFP, the ranking of the declared efficiencies is maintained for pLux, but not for pTet and J23101. The RBS B0030 results to be the most efficient for both J23101 and pTet, but not for pLux (NB: this effect might be due to an effective non-modularity of RBS, but also to saturating phenomena occurring for this very strong promoter at full induction). RBS B0031 always shows a very low efficiency, while B0032 an intermediate efficiency between B0031 and the stronger RBSs B0030 and B0034.
For what concerns the encoded gene variation, more significant differences can be observed. RBS B0030 has the higher efficiency only for mRFP, while the values for AiiA and LuxI are similar (~0.5). Unexpectedly, the weak RBS B0031 has a higher efficiency with AiiA gene (0.83), while with mRFP and LuxI. With B0032 no activity was observed for LuxI, while for AiiA and mRFP the results are quite consistent with the one reported above.
These results are encouraging: though the partial non-modularity of RBS with the encoded gene is confirmed, the hypothesis of modularity with the promoter is to some extent confirmed. Three classes of efficiencies were identified:
- low efficiency RBS (B0031)
- medium efficiency RBS (B0032)
- high efficiency RBSs (B0030, B0034)
pLux promoter
- BBa_K516330 (wiki name: E17 ) pLambda-RBS30-LuxR-T-pLux-RBS30-mRFP-TT
- BBa_K516331 (wiki name: E18 ) pLambda-RBS30-LuxR-T-pLux-RBS31-mRFP-TT
- BBa_K516332 (wiki name: E19 ) pLambda-RBS30-LuxR-T-pLux-RBS32-mRFP-TT
- BBa_K516334 (wiki name: E20 ) pLambda-RBS30-LuxR-T-pLux-RBS34-mRFP-TT
The estimated parameters for the Hill functions are summarized in the table below. For more details on parameter estimation, see the model section.
| RBS | αpLux [(AUr/min)/cell] | δpLux [-] | ηpLux [-] | kpLux [ng/ml] |
| BBa_B0030 | 438 [10] | 0.05 [>100] | 2 [47] | 1.88 [27] |
| BBa_B0031 | 9.8 [7] | 0.11 [57] | 1.2 [29] | 1.5 [26] |
| BBa_B0032 | 206 [3] | 0 [>>100] | 1.36 [10] | 1.87 [9] |
| BBa_B0034 | 1105 [6] | 0.02 [>100] | 1.33 [19] | 2.34 [18] |
From this table, it is evident that, whilst αpLux assumes significantly different values for different RBSs, ηpLux and kpLux assume very similar values. This result shows that RBS variation modulates the amplitude of Hill function, not affecting the shape of the curve. The four induction curves result to be the same Hill function modulated in amplitude by a parameter, that is the estimated RBS efficiency for this measurement system.
These results are quite encouraging, because suggest that, given the non-modular behavior of RBS dpending on the encoded gene, the RBS has a modular behaviour respect to the promoter.
The operative parameters are summarized in the table below:
| RBS | RPUmax | RPUmin | Switch point [nM] | Linear boundaries [MIN; MAX] [nM] |
| B0030 | 4.28 | 0.20 | 1.08 | [0.36; 3.27] |
| B0031 | 4.93 | 0.55 | 0.25 | [0.03; 2.30] |
| B0032 | 9.49 | 0.02 | 0.47 | [0.07; 3.07] |
| B0034 | 21.53 | 0.51 | 0.53 | [0.08; 3.77] |
These operative parameters provide useful information on the behavior of this 3OC6-HSL inducible device. RPUmax assumes very different values in terms of RPUs. This can't be explained by RBS modulation, since RPUs have been evaluated by normalizing Scell of pLux-RBSx for the one of J23101-RBSx. It is evident that some nonlinear effect on maximum strength, maybe due to saturation phenomena on protein expression, occur. The same RPUs should be observed for every RBS, since the normalization by the standard reference used for RPUs computation should eliminate the RBS contribute. Here different RPUs are observed, maybe due to nonlinear RBS behavior or to saturation phenomena occurring with this very strong promoter. The switch point and linear boundaries are quite constant in all the cases, showing that the linear region of this system is not affected by RBS changes.
pTet promoter
- BBa_K516230 (wiki name: E21 ) pTet-RBS30-mRFP-TT
- BBa_K516231 (wiki name: E22 ) pTet-RBS31-mRFP-TT
- BBa_K516232 (wiki name: E23 ) pTet-RBS32-mRFP-TT
- BBa_I13521 pTet-RBS34-mRFP-TT
The protocols for the characterization of pTet promoter are reported in the pTet measurement section.
The data collected from the mRFP measurement systems were processed as described in data analysis section. The induction curves were obtained by fitting a Hill function as described in modelling section and the estimated parameters for pTet are reported in the pictures and in table below.
This promoter is widely studied and characterized usually using the strong RBS BBa_B0034. Here we have characterized its transcriptional strength as a function of aTc induction (ng/ml) for different RBSs. Three different induction curves were obtained and are reported in figure:
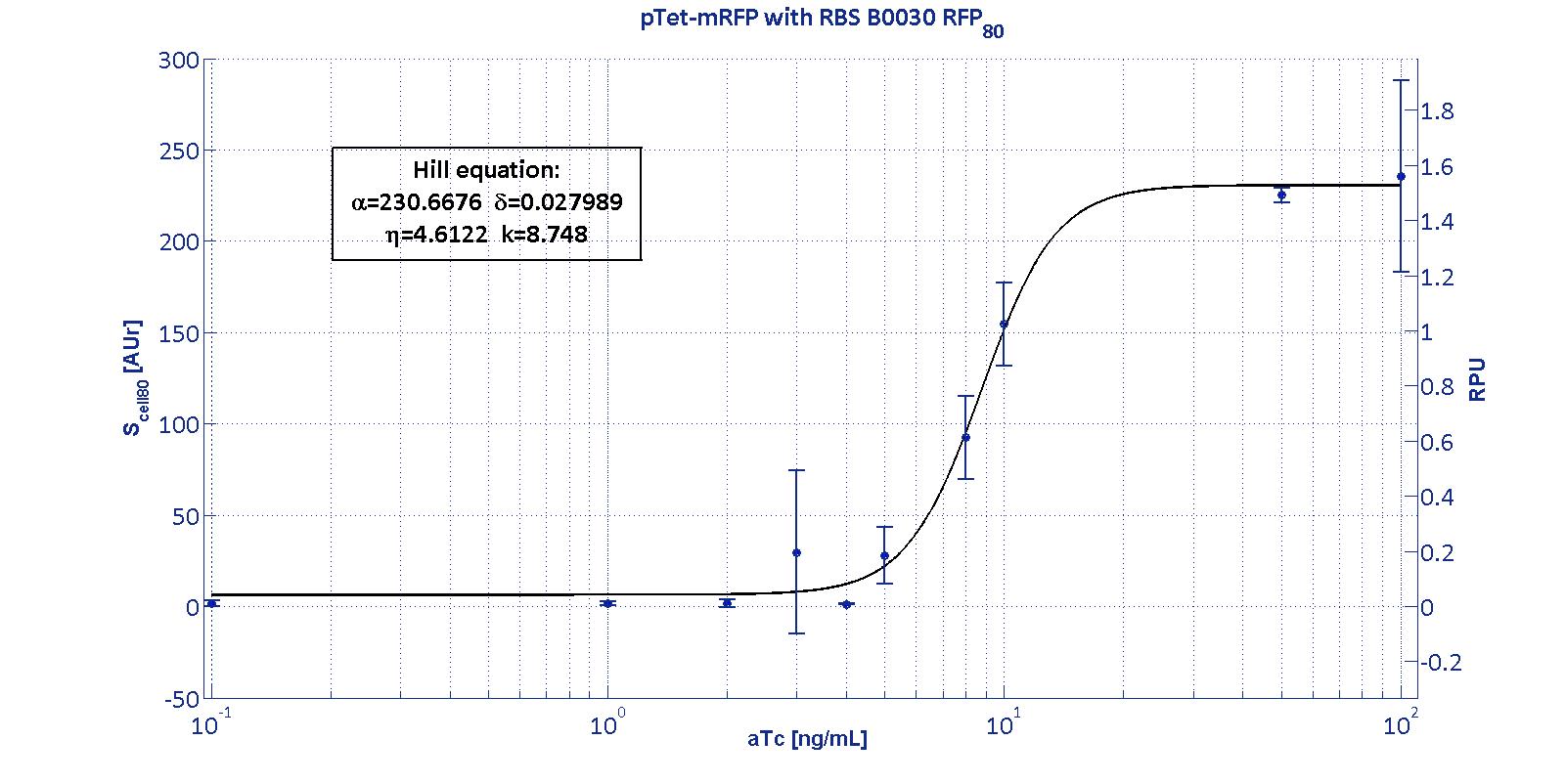
The estimated parameters of the Hill curves described in the figures are summarized in the table below:
| RBS | αpTet [(AUr/min)/cell] | δpTet [-] | ηpTet [-] | kpTet [nM] |
| BBa_B0030 | 230.67 [3.7] | 0.028 [91.61] | 4.61 [23.73] | 8.75 [4.16] |
| BBa_B0031 | ND | ND | ND | ND |
| BBa_B0032 | 55.77 [12] | 1.53E-11 [>>100] | 4.98 [57.62] | 7.26 [14.98] |
| BBa_B0034 | 120 [5.95] | 0.085 [40.6] | 24.85 [47.6] | 9 [5.43] |
The measurement system pTet-B0031-mRFP-TT couldn't be assayed because its fluorescence output is under the detectability threshold of our measurement instrument. For this reason, the parameters of the corresponding Hill curve couldn't be estimated and are reported as 'Not Determined' ND.
α parameter (representing the maximum trascriptional rate in the studied range of induction) varies as expected with the RBS variation and also the δ and η parameters are quite different among the RBS variations.
The kpTet parameter is quite constant among the RBS variations, thus suggesting that in this case the RBS variation doesn't substantially affect the switch point of the Hill curve, even if the amplitude and the maximum slope are not quite maintained (for the η parameter, maybe fitting problems).
The operative parameters are summarized in the table below:
| RBS | RPUmax | RPUmin | Switch point [ng/ml] | Linear boundaries [MIN; MAX] [ng/ml] |
| B0030 | 1.53 | ~0 | 7.95 | [4.66;11.99] |
| B0031 | ND | ND | ND | ND |
| B0032 | 3.16 | ~0 | 6.7 | [4.45;10.05] |
| B0034 | 2.73 | 0.23 | 8.96 | [8.27;9.71] |
From these parameters, it is evident that whilst the switch-point is almost maintained for all the RBSs, the linear boundaries are similar for RBS30 and RBS32 but for RBS34 are moved on the right of one order of magnitude.
LuxI gene - BBa_C0061
LuxI has been characterized through the Biosensor BBa_T9002 (see modelling section) using the pTet-RBSx-LuxI-TT measurement systems.
The HSL synthesis rate has been evaluated according to the model equations, properly adjusted. In particular, the ODE system is reported here:
Since the measurement systems are only assayed in the exponential growth phase, in the equation (3) N <
| Nmax | μ | γHSL |
| 1*109 | 0.004925 | 0 |
The parameters Vmax, kM,LuxI and αRBSx were estimated with a simultaneous fitting of the data collected as described in measurement section for the four measurement parts pTet-RBSx-LuxI-TT assayed by BBa_T9002 biosensor section.
The estimated parameters for the enzymatic activity of LuxI are reported in the table below:
| Vmax | kM,LuxI | αB0030 | αB0031 | αB0032 | αB0034 |
| 3.56*10-9 | 6.87*103 | 87 | 8.5 | ND | 252 |
The provided parameters kM and Vmax represent the enzymatic activity of LuxI, described by our model. They must not be confused with the operative parameters of the Michaelis-Menten relation. These synthetic parameters have a great importance, since they can be used in more complicated models in order to predict the behavior of complex circuits.
AiiA gene - BBa_C0060
The activity of AiiA enzyme has been evaluated by testing the measurement systems pTet-RBSx-AiiA-TT. Similar to LuxI, a system of differential equations (referring to model equations) has been derived.
The parameters kcat, kM,AiiA and αRBSx would have been estimated with a simultaneous fitting of the data collected as described in measurement section for the four measurement parts pTet-RBSx-AiiA-TT assayed by BBa_T9002 biosensor section.
Unfortunately, their estimation revealed difficult.
In the first experiments with the measurement system pTet-RBSx-AiiA-TT in LOW-COPY at pH=7 no degradation of HSL was observed. The collected data are shown in the figure below. HSL degradation is identical in the measurement system and in the negative control after 21 hours.
pSB1C3 plasmid with mRFP between S and P bearing pTet (easy-to-clone)
 |
Existing parts: sequence debugging
- BBa_K516021 (wiki name: E10 ) RBS31-AiiA-TT Rebuilt existing part from BBa_I13914 (DNA planning)
- BBa_K516022 (wiki name: E11 ) RBS32-AiiA-TT Rebuilt existing part from BBa_I13912 (DNA planning)
- BBa_K516030 (wiki name: E5 ) RBS30-mRFP-TT Rebuilt existing part from BBa_S04180 (DNA planning)
- BBa_K516032 (wiki name: E7 ) RBS32-mRFP-TT Rebuilt existing part from BBa_ J133000 (DNA planning)
- BBa_K516214 (wiki name: E16 ) pTet-RBS34-LuxI Rebuilt existing part from BBa_S03623 (DNA available, only 2008 kit, inconsistent)
- BBa_K516222 (wiki name: E26 ) pTet-RBS32-AiiA-TT Rebuilt existing part from BBa_ J22071 (2008 only, Bad sequencing)
- BBa_K516224 (wiki name: E27 ) pTet-RBS34-AiiA-TT Rebuilt existing part from BBa_K077047 (Part deleted)
- BBa_K516232 (wiki name: E23 ) pTet-RBS32-mRFP-TT Rebuilt existing part from BBa_I20252 (DNA planning)
 "
"