Team:Peking S/project/wire/matrix
From 2011.igem.org
Spring zhq (Talk | contribs) |
|||
| Line 39: | Line 39: | ||
[[File:SR_Matrix_table3.png|600px]]</center> | [[File:SR_Matrix_table3.png|600px]]</center> | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <a href="#top">Top↑ | ||
| + | </div> | ||
| + | |||
| + | </html> | ||
| + | <br> | ||
| + | <br> | ||
Latest revision as of 01:13, 6 October 2011
Template:Https://2011.igem.org/Team:Peking S/bannerhidden Template:Https://2011.igem.org/Team:Peking S/back2
Template:Https://2011.igem.org/Team:Peking S/bannerhidden

Chemical Wire Toolbox
Introduction|Harvesting ‘Chemical Wires’ From Nature|Synthesizing Quorum Sensing Inverters|Orthogonal Activating Matrix
Orthogonal activating matrix
Aiming at effective and discrete communications in a multicellular genetic network, orthogonality of the 'chemical wires' is essential. Thus we carried out a series of experiments checking the signaling orthogonality of 'chemical wires' in our toolbox.
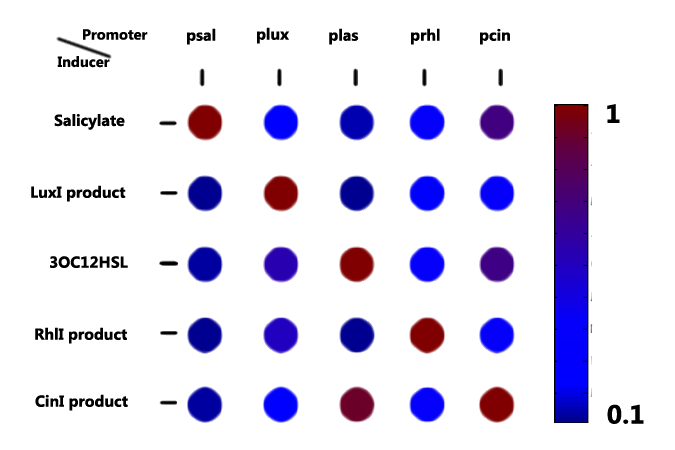
Figure 1 A graphical representation of the relative activation fold associated with five receiver systems and their corresponding inducer.The concentration of salicylate is 10^-4 mol/L. LuxI product and RhlI product is 10 fold dilution of the supernatant from a overnight culture bearing pT7 controlled luxI generator and ptet controlled rhlI generator, respectively. The concentration of 3-oxododecyl-homoserine lactone(3OC12HSL) is 10^-8 mol/L, and the CinI product is 1:100 dilution of the overnight culture broth.
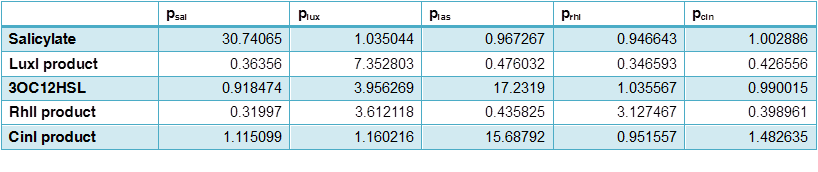
Table 2 Data of the activating matrix, average fold compared with blank samples calculated
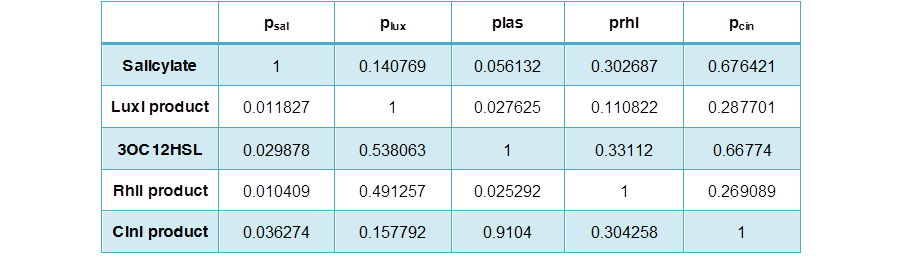
Table 3 Data of the activating matrix, activating fold normalized
 "
"