Team:Peking S/project/wire
From 2011.igem.org
Nuanuan128 (Talk | contribs) |
Nuanuan128 (Talk | contribs) (→Reference) |
||
| (4 intermediate revisions not shown) | |||
| Line 29: | Line 29: | ||
</center> | </center> | ||
| - | For more details,[https://2011.igem.org/Team:Peking_S/project/wire/harvest click here] | + | ''For more details'', [https://2011.igem.org/Team:Peking_S/project/wire/harvest ''click here''] |
| Line 36: | Line 36: | ||
<center>[[File:NN10.png|400px]]</center> | <center>[[File:NN10.png|400px]]</center> | ||
| - | For more details, [https://2011.igem.org/Team:Peking_S/project/wire/inverter click here] | + | ''For more details'', [https://2011.igem.org/Team:Peking_S/project/wire/inverter ''click here''] |
| Line 43: | Line 43: | ||
<center>[[File:signalingOrthogonal.png|450px]]</center> | <center>[[File:signalingOrthogonal.png|450px]]</center> | ||
| - | For more details,[https://2011.igem.org/Team:Peking_S/project/wire/matrix click here] | + | ''For more details'', [https://2011.igem.org/Team:Peking_S/project/wire/matrix ''click here''] |
| + | |||
| + | |||
| + | == Reference == | ||
| + | |||
| + | |||
| + | [1] Li, B. You, L. Synthetic biology: Division of logic labour. Nature 469, 171–172 (2011). | ||
| + | |||
| + | [2] Marguet, P. Balagadde F. Tan C. You L. Biology by design: reduction and synthesis of cellular components and behaviour. JR Soc Interface 4, 607–623 (2007). | ||
| + | |||
| + | [3] Voigt, C.A. Genetic parts to program bacteria. Curr Opin Biotechnol 17, 548–557 (2006). | ||
| + | |||
| + | [4] Clancy, K, Voigt, C.A. Programming cells: towards an automated 'Genetic Compiler'. Curr Opin Biotechnol 21, 572–581 (2010) . | ||
| + | |||
| + | |||
<html> | <html> | ||
<a href="#top">Top↑ | <a href="#top">Top↑ | ||
</div> | </div> | ||
| + | |||
</html> | </html> | ||
<br> | <br> | ||
<br> | <br> | ||
Latest revision as of 03:54, 6 October 2011
Template:Https://2011.igem.org/Team:Peking S/bannerhidden Template:Https://2011.igem.org/Team:Peking S/back2
Template:Https://2011.igem.org/Team:Peking S/bannerhidden

Chemical Wire Toolbox
Introduction|Harvesting ‘Chemical Wires’ From Nature|Synthesizing Quorum Sensing Inverters|Orthogonal Activating Matrix
Introduction to ‘Chemical Wire’ toolbox
Cell-cell communication-based multicellular networks provide an extended vista for synthetic biology. By compartmentalizing complex genetic circuits into separate engineered cells, the difficulty of the construction by layering elementary gates can be dramatically reduced, partly due to the insulation of crosstalk between modules, the suppression of noise by populationally averaging, and the reducing of metabolic burden in host cells. What’s more, cell-cell communication-based multicellular feature enables coordination and synchronization among cells in and between populations and facilitates the generation of reliable non-Boolean dynamics.
However, orthogonal ‘chemical wires’ that allow concurrent communication are far from sufficient, making developing a versatile ‘chemical wire’ toolbox for conducting a complex gene network more necessary. This year we are aiming to develop a ‘chemical wire’ toolbox, applicable for both Boolean and Non-Boolean gene networks.
As natural quorum sensing systems provide an excellent pool for developing ‘chemical wire’ toolbox, harvesting ‘chemical wires’ from the nature is a fast and affordable way. We selected and evaluated a recently reported quorum sensing system, CinI-CinR system from Rhizobium leguminosarum as a candidate of our toolbox. An artificial QS system, PchAB-NahR system exploiting salicylate as signaling molecule was also built. Both of them were proved to owe promising performance.
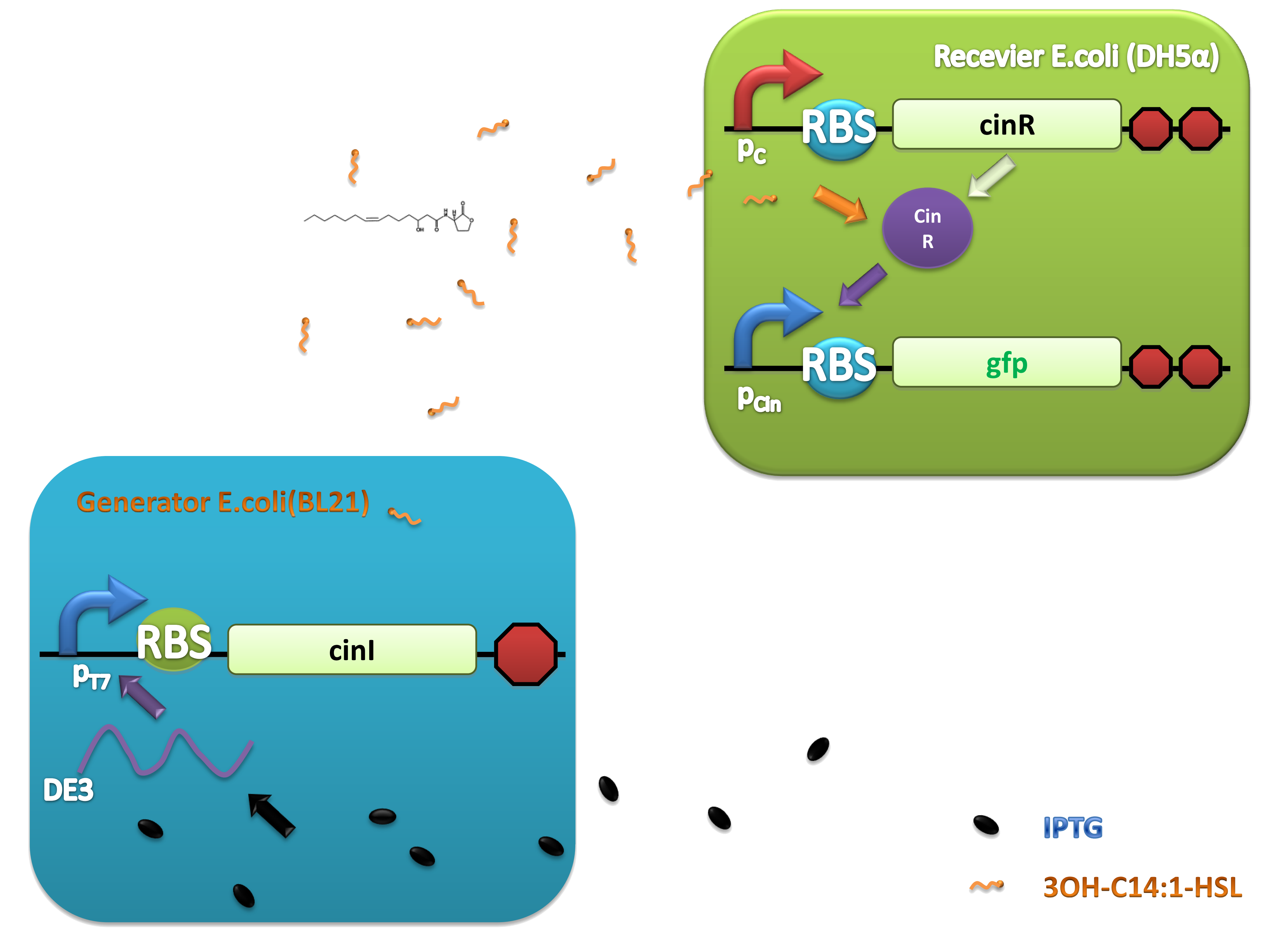
For more details, click here
On the other hand, all of the quorum sensing systems currently exploited in synthetic biology exhibit transcriptional activation, which cannot provide negative feed back loops during cell-cell communication, and the conventional inverter, which was implemented by the repressor-operator pairs, has evident defects. We have developed a ‘from ground up’ approach to synthesize direct, fast and reliable signaling inverters for synthetic microbial consortia, harnessing the conditioned binding of quorum sensing regulators to their cognate DNA boxes to control the accessibility of RNA polymerase.
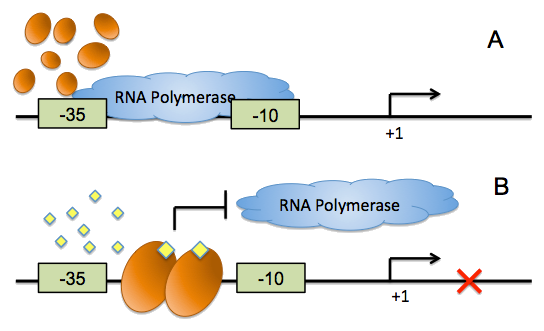
For more details, click here
With candidates harvested from nature or re-designed from the natural counterpart, we started to characterize them, specially focusing on their orthogonality, dose response, time dependence (signaling speed) and their ability to coordinate cell population.
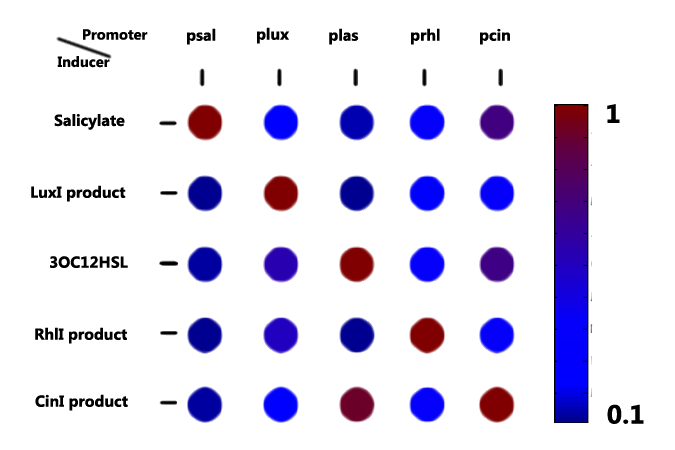
For more details, click here
Reference
[1] Li, B. You, L. Synthetic biology: Division of logic labour. Nature 469, 171–172 (2011).
[2] Marguet, P. Balagadde F. Tan C. You L. Biology by design: reduction and synthesis of cellular components and behaviour. JR Soc Interface 4, 607–623 (2007).
[3] Voigt, C.A. Genetic parts to program bacteria. Curr Opin Biotechnol 17, 548–557 (2006).
[4] Clancy, K, Voigt, C.A. Programming cells: towards an automated 'Genetic Compiler'. Curr Opin Biotechnol 21, 572–581 (2010) .
 "
"