Team:MIT/Notebook/
From 2011.igem.org
Overview
Here you can see what we did weekly in the lab!
Week 1: June 6 - June 11
In the first week we began constructing basic reporter DNA constructs that have various combinations of promoters and genes. The mix-and-matching of promoters and genes was done using the LR reaction following the Gateway protocol. Our promoters included TRE, minCMV, Hef1a, Hefl1a-LacO, and our genes included eYFP, mKate, and eBFP2. Transformed and inoculated colonies from the LR reactions and performed restriction digests. Approximately half of the LR reactions were successful and became DNA parts that we could use later on.June 7, 2011
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Tube Number |
Colony Number, Antibiotic, Person |
Protocol Used |
|---|---|---|---|---|---|---|---|---|
| 6.7.2011 |
Michelle Louis |
2-3 |
Tre |
C1434VP16 |
Yes. 6.7.2011, 5pm |
11 |
5, AMP, KH |
Standard, 30C for 16h |
| 6.7.2011 |
Michelle Louis |
2-3 |
Tre |
Mnt1VP16 |
Yes. 6.7.2011, 5pm |
12 |
4, AMP, KH | Standard, 30C for 16h |
| 6.7.2011 |
Tyler |
2-3 |
Tre |
LacIKrab |
Yes. 6.7.2011, 5pm |
6 |
30, AMP, KH | Standard, 30C for 16h |
| 6.7.2011 |
Jenny Divya |
3-4 |
minCMV-7xMnt1 |
eYFP |
Yes. 6.7.2011, 5pm |
9 |
0, AMP, KH, needs to be redone |
Standard, 30C for 16h |
| 6.7.2011 |
Jenny Divya |
2-3 |
Tre |
LexAVP16 |
Yes. 6.7.2011, 5pm |
10 |
2, AMP, KH, needs to be redone |
Standard, 30C for 16h |
| 6.7.2011 | Mariola Kenneth |
3-4 |
minCMV 1xC1434 |
eYFP |
Yes. 6.7.2011, 5pm |
7 |
3, AMP, KH, needs to be redone | Standard, 30C for 16h |
| 6.7.2011 | Mariola Kenneth |
3-4 | minCMV 4xLexA |
eYFP |
Yes. 6.7.2011, 5pm |
8 |
0, AMP, KH, needs to be redone |
Standard, 30C for 16h |
LR Reactions
Utilizing protocol here: LR Protocol
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Tube Number |
Colony Number, Antibiotic, Person |
Protocol Used |
|---|---|---|---|---|---|---|---|---|
| 6.9.2011 |
Grant |
3-4 |
minCMV 4xLexA | eYFP | Yes. 6.9.2011, 3pm |
12 |
10, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
minCMV-4xMnt1 |
eYFP |
Yes. 6.9.2011, 3:45pm | 2 |
13, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
minCMV-4xCI434 |
eYFP |
Yes. 6.9.2011, 3:45pm | 1 |
7, AMP, GR | Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
Hef1a-LacO |
eYFP |
Yes, 6.9.2011, 4:20pm |
5 |
25, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Semon |
4-5 |
Tre |
mKate |
Yes, 6.9.2011 4:27pm |
3 |
6, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Semon |
1-2 |
Hef1a |
eBFP2 |
Yes, 6.9.2011, 4.27pm |
4 |
3, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
Transformation
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Colony Number, Antibiotic |
Protocol Used | |
|---|---|---|---|---|---|---|---|---|
| 6.9.2011 |
Mariola |
|
|
eYFP |
no. Replication stock. | |
2, AMP |
Transformation (~ 2 hr 20 min) using LB instead of SOC |
Miniprep
| Date | Assignee |
DNA |
Quantity |
Time collected |
|---|---|---|---|---|
| 6.9.2011 |
Divya-Jenny |
pEXPR_2-3_Tre:LexAVP16 |
52.2 ng/uL |
12pm |
| 6.9.2011 |
Kenneth |
pEXPR_3-4_minCMV-CI434:eYFP (A) |
70 ng/ul |
12pm |
| 6.9.2011 |
Kenneth | pEXPR_3-4_minCMV-CI434:eYFP (B) |
174 ng/uL |
12pm |
| 6.9.2011 |
Louis |
pEXPR_2-3_Tre:C1434VP16 |
234.6 ng/uL |
12pm |
| 6.9.2011 |
Tyler |
pEXPR_2-3_Tre:Lac/Krab |
130.7 ng/uL |
12pm |
| 6.9.2011 |
Michelle |
pEXPR_2-3_Tre:Mnt1VP16 |
110.0 ng/uL |
12pm |
| 6.9.2011 |
Michelle |
pDEST_2-3_ccdB |
117.6 ng/uL |
12pm |
Restriction Digests
| Assignee |
DNA |
Enzyme |
Expected Results |
Picture of Gel |
Time Incubated | Comments |
||
|---|---|---|---|---|---|---|---|---|
| Louis |
pEXPR_2-3_Tre:C1434VP16 | NdeI |
6700 bp 800 bp |
a.3 | 6/9/11 3:50 PM |
|
||
| Kenneth |
pEXPR_3-4_minCMV-CI434:eYFP (A+B) |
NcoI and SacII |
2450 bp 4650 bp |
A: a.6 B: a.7 |
6/9/11 3:00 PM |
A did not cut as expected. B looks good. Digestion Mix: 2 uL NEB4, 0.5 uL of each enzyme, A: 7.1 uL/B: 2.9 uL of DNA, 10.4 uL/14.6 uL of H20 |
||
| Michelle |
pEXPR_2-3_Tre:Mnt1VP16 |
BglI |
3700 bp 2200 bp 1300 bp |
a.1 | 6/9/11 4:00PM |
DNA appeared to be uncut. Need to redo, possibly with a new enzyme and more DNA. |
||
| Michelle |
pDEST_2-3_ccdB |
NcoI and NheI |
6100 bp 1700 bp |
a.2 | 6/9/2011 4:25 PM |
Attained bands at correct positions. Other band represents partially cut DNA in double digest. |
||
| Divya |
pEXPR_2-3_Tre:LexAVP16 |
HincII |
a.4 | 6/9/2011 4:45 PM |
- Bands didn't show up. Needs to be redone. - DNA may have degraded. Need to redo Nanodrop as well. |
|||
| Tyler | pEXPR_2-3_Tre:Lac/Krab | SalI | 3 bands: 5288 bp 1510 bp 1241 bp cuts gene |
a.5 | 6/9/2011 3:00 PM |
Digestion: 4 uL * 130.7 ng/uL = 522.8 ng DNA 2 uL NEB3 2 uL BSA 11 uL H2O 1 uL SalI Total: 20 uL Only saw one band at 3500 bp Needs to be redone possibly with another restriction enzyme |
Gels
June 10, 2011
Restriction Gels
| Assignee | DNA |
Enzyme |
Expected result |
Time Incubated |
Comments |
|---|---|---|---|---|---|
| Louis |
pEXPR_2-3_Tre:C1434VP16 | EcoRI (Buffer 2) |
5500 bp 1050 bp 650 bp 360 bp |
6/10/11 11:15 AM |
|
| Michelle |
pEXPR_2-3_TRE:Mnt1VP16 (A) |
NcoI and NheI |
5700 bp 1300 bp |
6/10/11 11:40 AM |
Digestion: 6 uL DNA 1 uL NcoI 1 uL Nhe1 2 uL NEB2 2 uL BSA 8 uL H2O Failed. |
| Michelle |
pEXPR_2-3_TRE:Mnt1VP16 (B) |
NheI and SphI |
1700 bp 5300 bp |
6/10/11 11:40 AM |
Digestion: 6 uL DNA 1 uL NheI 1 uL SphI 2 uL NEB2 2 uL BSA 8 uL H2O Failed. Redoing LR on Monday June 13th. |
| Tyler | pEXPR_2-3_Tre:Lac/Krab | SalI | 3 bands: 5288 bp 1510 bp 1241 bp cuts gene |
6/10/2011 3:00 PM |
Digestion: 4 uL * 130.7 ng/uL = 522.8 ng DNA 2 uL NEB3 2 uL BSA 11 uL H2O 1 uL SalI Total: 20 uL ***Worked (see gel below - b.1) Stored in -80C freezer |
Comments:
pEXPR_2-3_Tre:LexAVP16 and pEXPR_3-4_minCMV-7xMnt1:eYFP are being entirely redone.
pEXPR_2-3_Tre:LexAVP16 (post-miniprep) was lost. pEXPR_3-4_minCMV-7xMnt1:eYFP LR was unsuccessful (no bacteria grew).
LR and transformation will be done on Saturday. Miniprep and restriction mapping will be done on Sunday
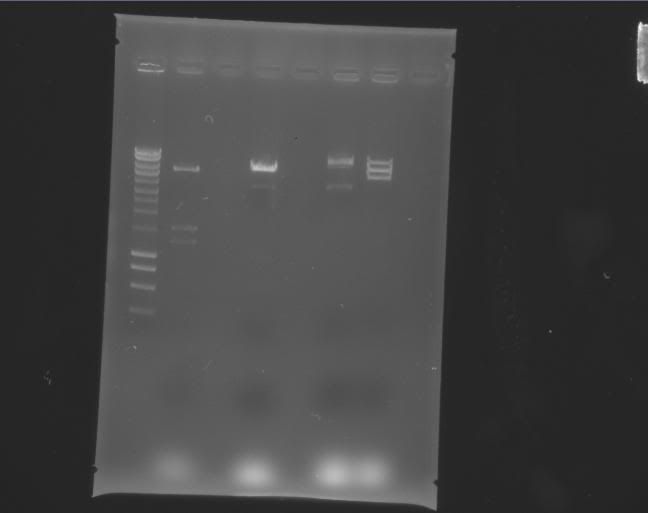
| Label | Gel |
Legend |
|---|---|---|
| b |
 > > |
Column 0: HyperLadder Column 1: pEXPR_2-3_Tre:Lac/Krab Column 4: pEXPR_2-3_Tre:C1434VP16 Column 6: pEXPR_2-3_TRE:Mnt1VP16 (A) Column 7: pEXPR_2-3_TRE:Mnt1VP16 (B) |
June 11, 2011
Set of LRs from 6/9 (GR, JC, and SR) cell counts taken; inoculated by GR/JC. 3 cells taken per plate.
| Assignee |
DNA |
Enzyme |
Expected Results |
Time Incubated |
Comments |
|---|---|---|---|---|---|
| Sam |
pEXPR_1-2_Hef:eBFP |
NcoI & ApaL1 |
3750bp, 3180bp, 1250bp | 45 min | If it doesn't work there should be a whole mess of fragments (including a small 600bp one) 1mL each enz; 2 mL Buf; 1 mL BSA; 2mL DNA; 13mL H2O (Standard Protocol with extra BSA) |
| Sam |
pEXPR_4-5_Tre:mKate |
Bgl1 |
1270bp, 2630bp, 3380bp | 45 min | 1mL enz; 2mL DNA; 2mL BSA; 2mL Buf; 15mL H2O (Standard Protocol with extra BSA and water - not quite 10x) |
 "
"