Team:Kyoto/Digestion
From 2011.igem.org
(→3,5-dinitorosalicylic acid assay(DNS method)) |
(→preliminary experiments) |
||
| Line 90: | Line 90: | ||
#two, three, five hours after, we did above operation, taking supernatant, measured OD500, heating and cooling, applying DNS reagent and heating and cooling again. | #two, three, five hours after, we did above operation, taking supernatant, measured OD500, heating and cooling, applying DNS reagent and heating and cooling again. | ||
#added all sample tube (containing 320㎕ solution) distilled water by 2ml and measure the absorbance of them in 550nm. | #added all sample tube (containing 320㎕ solution) distilled water by 2ml and measure the absorbance of them in 550nm. | ||
| + | |||
| + | [[File:キチナーゼ.PNG]] | ||
== '''Result''''' == | == '''Result''''' == | ||
Revision as of 21:34, 4 October 2011
Contents |
Project Digestion
Introduction
Streptomyces is a kind of prokaryotic bacteria which decompose bodies in nature[1]. We extract chitinase gene from this bacterium and introduce into Escherichia coli. Secretion-signal sequences are included in this gene so that the protein coded by them will go out without occurring cell lysis. After assembling all genes, we examined the activity of these two enzymes in both of and quantitative ways.
Method
Construction
We created following constructions to allow secretion of chitinase, chiA1. This gene is regulated by lactose promoter, BBa_R0011. We used Streptmyces’s RBS into this constructions, because reference article [1] used that to allow E.coli to secrete the protein.
Assay
We performed following qualitative and quantitative assays. This method take a little time and cost a little money.
3,5-Dinitrosalicylic acid assay (DNS method)
- This assay is based on this fact: 3,5-dinitorosalicylic acid (DNS) is changed into 3-amino- 5-nitorosalicylic acid by reducing saccharide in reaction solution and the absorbance of this liquid increase in direct proportion to the amount of reducing sugar.
- If chitinase is secreted, chitin in media was decomposed to, for example, N-acetylglucosamin,that is ,reducing sugar. Therefore, The absorbance of the media added DNS will increase.
This assay is based on this fact: 3,5-dinitorosalicylic acid (DNS) is changed into 3-amino- 5-nitorosalicylic acid by reducing saccharide in reaction solution and the absorbance of this liquid increase in direct proportion to the amount of reducing sugar.
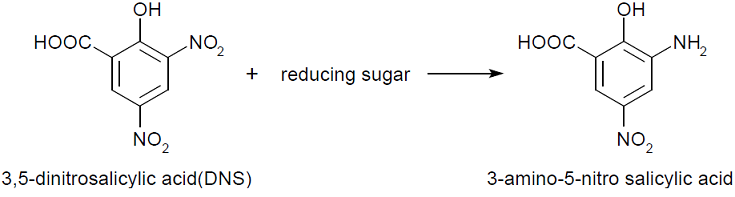
To carry out this assay, we will be required following three things, that is,
- the relationsip to reducing sugar concntration and absorbance 550
- the assesment of effect of E.coli in media
- the effect of chitinase
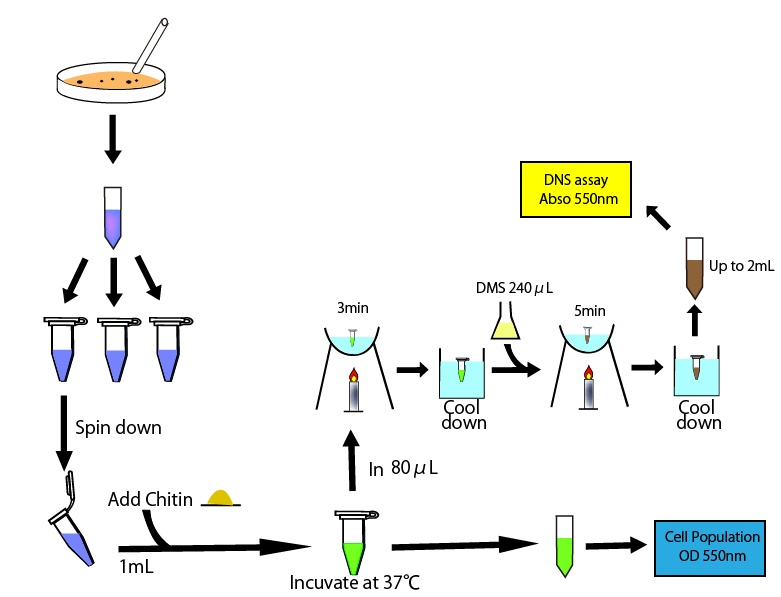
Method
Procedure
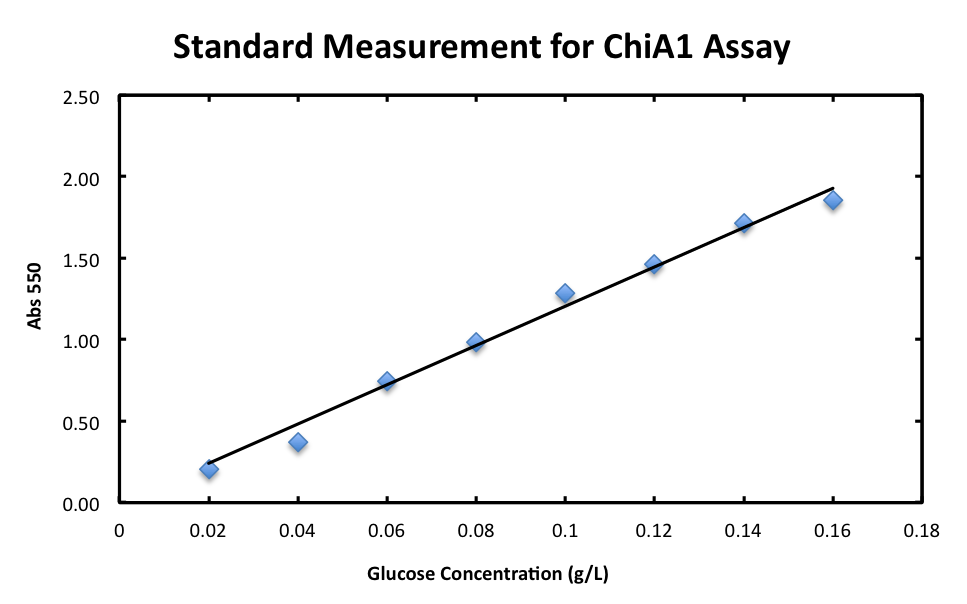
DNS method We examined the quantitative relation between absorbance and the volume of sugar and then expressed it onto a straight line graph (result fig 1:リンク).
We led E.coli introduced chitinase gene had secreted disassemble chitin into N-acetylglucosamine and other sugar derivatives in water. After passing enough time (_min), we added this liquid 0.2 ml into DNS reagent 0.6 ml (How to prepare) and boiled this solution for 5 min. After cooling this soluion, We diluted with water to 5 ml and assay the absorbance in 550 nm.
The results of this measurement and the fig 1 graph enabled us to calculate the amount of digested chitin, showing the relative activity of chitinase.
preliminary experiments
1-1:evaluate the affection of medium
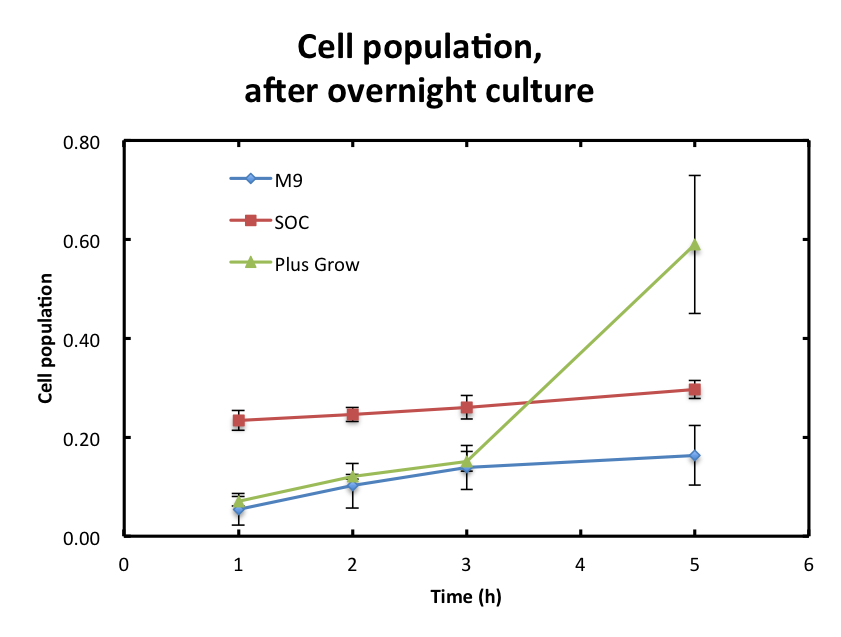
we examined the interruption based on the components of media(SOC, plas-grow and M9)
| Sample | glucose solution |
| Blank | mixed liquid (240μℓDNS reagent plus 1760㎕distilled water ) |
| Reagent | DNS |
- added 240㎕ DNS reagent to 80㎕sample
- heated it for 5 min in boiling water and then cooled it in water.
- added it distilled water by 2ml
- measured absorbance in 550nm
1-2:measurement the effect of remainded E.coli
This assay was performed three times for each medium
- poured the medium cultured E.coli overnight 1.2 ㎕ into each five microcentritube.
- centrifuged them for 5 min at 5,000 rpm
- prepared new five microcentritube and move 800㎕ the supernatants into each of them.
- measure the OD550 of one tube(use fresh medium as a blank in following assays)
- one hour after, we measured OD 550 of other tube
- take 80㎕ supernatant and move it into new tube and then heated it for 3 min in boiling water and then cooled it in water.
- added 240㎕ DNS reagent and heated it for 3 min in boiling water and then cooled it in water.
- two, three, five hours after, we did above operation, taking supernatant, measured OD500, heating and cooling, applying DNS reagent and heating and cooling again.
- added all sample tube (containing 320㎕ solution) distilled water by 2ml and measure the absorbance of them in 550nm.
Result
1. Standard Measurement for ChiA1.
- From the result, a strong correlation between glucose concentration and its A550 was observed.
2. Consideration of medium and growth of E.coli.
Discussion
From result of preliminary experiments, we found several problems. ・the affection of medium itself
The components of each medium also reduced 3,5-dinitorosalicylic acid and would cause error in the assay.
・interruption of remaining E.coli.
Even though we use the centrifugal supernatant, there was still some E.coli. we found they could interrupt data because they would decompose reducing sugers.
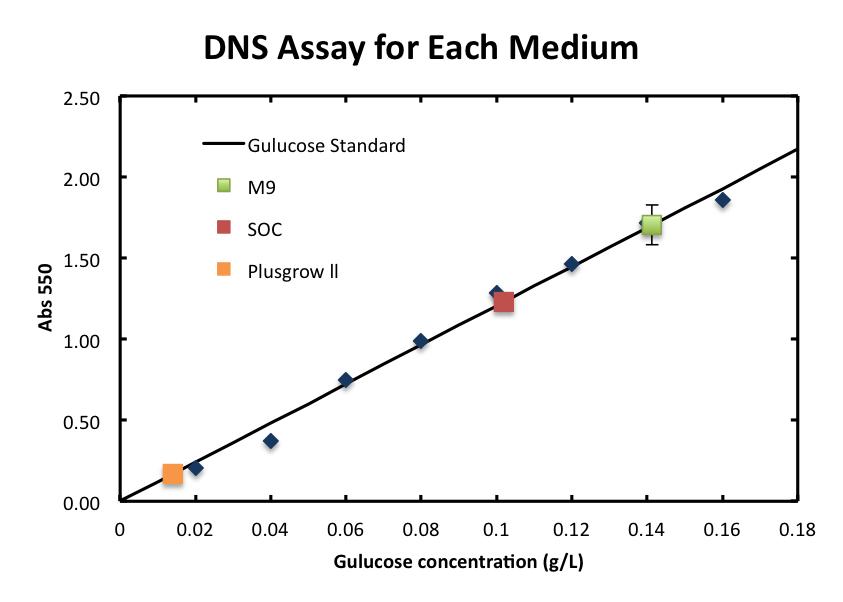
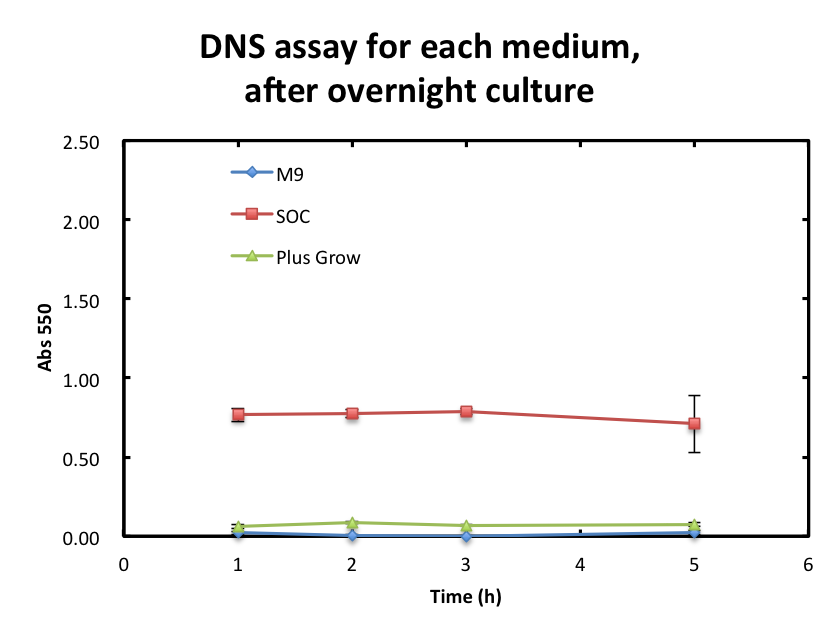
To overcome these barriers, we decided detail plan of our assay. From the result fig 3, SOC medium cultured E.coli overnight would still include too much amount of reducing materials and, from fig 3. plas-grow enabled reminded E.coli to grow rapidly. However, as for M9 medium,
Result
1. Standard Measurement for ChiA1.
- From the result, a strong correlation between glucose concentration and its A550 was observed.
2. Consideration of medium and growth of E.coli.
- We checked the influence of each medium to the DNS assay. Figure 2 shows the background absorbance of each medium. The absorbance of M9 was 1.7±0.1, SOC was1.227±0.007, and Plusgrow Ⅱ was 0.17±0.02.
Discussion
From result of preliminary experiments, we found several problems.
・the affection of medium itself
The components of each medium also reduced 3,5-dinitorosalicylic acid and would cause error in the assay.
・interruption of remaining E.coli.
Even though we use the centrifugal supernatant, there was still some E.coli. we found they could interrupt data because they would decompose reducing sugers.
To overcome these barriers, we decided detail plan of our assay.
From the result fig 3, SOC medium cultured E.coli overnight would still include too much amount of reducing materials and, from fig 3. plas-grow enabled reminded E.coli to grow rapidly. However, as for M9 medium,
Reference
[1] “Actinobacteria.” Internet: http://en.wikipedia.org/wiki/Actinobacteria [Nov. 5, 2011]
[2] A. S. Khalil, J. J. Collins, “Synthetic biology: applications come of age.” Nat Rev Genet., vol. 11, no. 5 pp. 367-79, May. 2010.
[3] H. Ikeda, J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, S. Omura, “Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis.” Nat Biotechnol., vol. 21, no. 5 pp. 526-31, Apr. 2003
[4] S. Omura, H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, “Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites.” Proc Natl Acad Sci U S A. vol. 98, no. 21 pp. 12215-20, Oct. 9
[5] H. Sakuzou, ”還元糖の定量法(生物化学実験法)” Kyoto University: Japan Scientific Soceties Press
[6] S. Kongruang, M. J. Han, C. I. Breton, M. H. Penner, “Quantitative Analysis of Cellulose-Reducing Ends.” Appl Biochem Biotechnol. Vol. 113, no. 116 pp. 213-31, Spring 2004
 "
"