Team:ITESM Mexico
From 2011.igem.org
| Line 1: | Line 1: | ||
{{Team:ITESM_Mexico/Top}} | {{Team:ITESM_Mexico/Top}} | ||
[[File:Sensecoli-resumen.png|left]] | [[File:Sensecoli-resumen.png|left]] | ||
| - | + | Using previously developed mechanisms, this project will develop a new system capable of detecting a desired analyte, the system will report the concentration of such analyte at a given time lapse with the possibility of setting an instant concentration reading at will. Since most of the mechanisms used are based in detecting arabinose, this project will use arabinose as the measured analyte to avoid unnecessary difficulties. | |
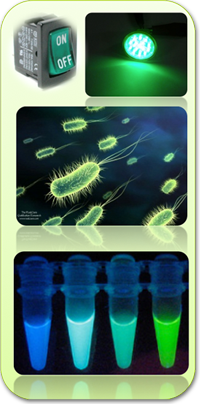
| - | + | Our project is a biosensor for arabinose build in an E. coli chassis, it will measure the concentration levels of arabinose by reporting two different fluorescent proteins, GFP for a low concentration level and CFP for a high concentration level. | |
| - | + | Based on the work of the Tokyo-NokoGen (2010) project, Chiba University (2010), UNAM-Genomics México (2010), British Columbia (2009), Cambridge (2010) and ITESM campus Monterrey (2010) we fused two ideas: creating a sensor that will measure the concentration level at out command and it will be reset-able. This will allow us to obtain a concentration reading of arabinose whenever we want it, improving the current lock-and-key mechanism. It will also include a master switch mechanism that works with a biological photosensor included in our design. Exposing our bacterial culture to green light will activate this mechanism, switching the biosensor on. | |
| - | The results will be obtained by reading the amount of light with a flux-cytometer. This way we will not only obtain a concentration range but a precise concentration reading. | + | The aim of this project is to design three bimolecular mechanisms that will enable the organism to 1) Detect the presence of a desired analyte (arabinose) and give a color response accordingly to its concentration. 2) Determine the concentration at regular time intervals 3) Respond to a non-chemical signal to give an immediate reading of the concentration. |
| - | + | ||
| + | The results will be obtained by reading the amount of light with a flux-cytometer. This way we will not only obtain a concentration range but a precise concentration reading. | ||
| + | |||
| + | In the future this biosensor will work as a chassis to detect a wide variety of compounds such as toxins and/or antibodies. | ||
Revision as of 22:15, 14 July 2011
Using previously developed mechanisms, this project will develop a new system capable of detecting a desired analyte, the system will report the concentration of such analyte at a given time lapse with the possibility of setting an instant concentration reading at will. Since most of the mechanisms used are based in detecting arabinose, this project will use arabinose as the measured analyte to avoid unnecessary difficulties.
Our project is a biosensor for arabinose build in an E. coli chassis, it will measure the concentration levels of arabinose by reporting two different fluorescent proteins, GFP for a low concentration level and CFP for a high concentration level.
Based on the work of the Tokyo-NokoGen (2010) project, Chiba University (2010), UNAM-Genomics México (2010), British Columbia (2009), Cambridge (2010) and ITESM campus Monterrey (2010) we fused two ideas: creating a sensor that will measure the concentration level at out command and it will be reset-able. This will allow us to obtain a concentration reading of arabinose whenever we want it, improving the current lock-and-key mechanism. It will also include a master switch mechanism that works with a biological photosensor included in our design. Exposing our bacterial culture to green light will activate this mechanism, switching the biosensor on.
The aim of this project is to design three bimolecular mechanisms that will enable the organism to 1) Detect the presence of a desired analyte (arabinose) and give a color response accordingly to its concentration. 2) Determine the concentration at regular time intervals 3) Respond to a non-chemical signal to give an immediate reading of the concentration.
The results will be obtained by reading the amount of light with a flux-cytometer. This way we will not only obtain a concentration range but a precise concentration reading.
In the future this biosensor will work as a chassis to detect a wide variety of compounds such as toxins and/or antibodies.
 "
"