Team:HokkaidoU Japan/Project/T3SS
From 2011.igem.org
HokkaidoU Japan
iGEM 2011 Team of Hokkaido University
Contents |
- Abstract
- What`s T3SSDetailed information about T3SS and summary of our achievements on iGEM 2010
- Injection assay using onion cellsExperiments using plant cells are easier to perform than with mammalian ones
- Ready-to-inject backbone and Bsa I cloning siteReady-to-inject backbone and Bsa I cloning site enables easy fusion of T3S signal and protein
- GSK tag systemA neat injection assay using GSK tag, which can specifically detect successfully injected proteins
- Bsa I cloning site, RFC submissionDetailed documentation of costructing a BioBrick cloning site a BioBrick!
T3SS
Introduction

Type III Secretion System (T3SS) is a system of pathogenic gram-negative bacterium such as Salmonella, Yersinia and EPEC(entero pathogenic E. coli). Using this system bacteria can inject whole protein molecules through a syringe like organelle named Type 3 Secretion Apparatus (Fig. 1). The target of this system is a eukaryotic cell. Naturally it is used to inject Virulence effector proteins.
Structure of Type III secretion apparatus
Type III secretion apparatus have a syringe like structure (Fig. 1). It can be visible under electron microscope [1]. Its length is about 80 nm and the diameter of its needle channel is about 2 nm [2]. The length is about 1/10 and the diameter is about 1/400 of an E. coli cell’s minor axis.
How does it function?
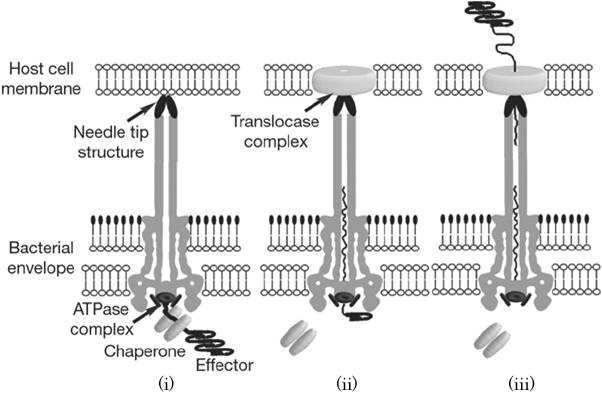
When the needle tip attaches to a target cell membrane, a translocator complex is assembled on the target cell membrane. Inside the bacterial cell, an effector protein which has a unique secretion signal domain on its N-terminal is recognized by the specific chaperone, and forms effector-chaperone complex. This complex is recognized by the T3-secretion-associated ATPase located at the base of the needle complex (Fig. 2 i). Then, the ATPase stripes the chaperone from the complex and unfolds the effector protein to pass through the channel of the needle complex (Fig. 2 ii). Finally, the translocated effector is refolded within the target cell to carry out its function (Fig. 2 iii)[1].
Our achievements on iGEM 2010
We made a T3SS test construct on pSB1T3 vector, and transformed it to the E. coli (K-12) carrying BAC vector "pBAC-SPI-2" that encodes a part of Salmonella enterica serovar Typhimurium LT2 genome, which would express secretion apparatus (Fig. 3).
Secretion signal
It was reported that N-terminus 191 amino acid residues of SlrP (one of the natural effector protein of SPI-2 encoded T3SS) function as a T3 secretion signal domain in Salmonella[3].
NLS (Nuclear Localization Signal)
In addition to the signal peptide, we located triple NLS (Nuclear Localization Signal) repeats between the T3 secretion signal and GFP, so that the injected GFP would be localized to the nucleus in the target cell.
RFP reporter
The RFP reporter gene constitutively produces RFP as an internal control that cannot be injected into target cells. This RFP would also play a role to distinguish GFP located in target cells from GFP remaining in bacterial cells under the fluorescence microscope.
Arabinose promoter
This [Secretion signal-NLS-GFP] fusion gene is under control of inducible arabinose promoter, this E. coli produces GFP only in the presence of arabinose.
Methods of Infection Assay
We transferred E. coli [SPI-2/ T3 Signal-GFP, RFP] overnight LB culture to the neutral pH Mg2+ minimal medium (MgM pH=7.2) and cultured for 4 hours with arabinose to accumulate sufficient amount of GFP fusion protein (Fig. 4 i). Then the bacteria were transferred to acidic pH Mg2+ minimal medium (MgM pH=5.0) and cultured for 4 hours with arabinose to assemble T3 secretion apparatus[4] (Fig. 4 ii). After that, bacteria were washed three times to remove contaminants that may detrimental for target cells (Fig. 4 iii). Since T3SS encoded in SPI-2 is known to be function inside of the phagosome of target cells[5], the bacteria were exposed to the cells (RK13) under acidic pH condition (pH=5.0), which is considered to be similar to the environment inside of the phagosome.
Under the acidic pH condition after the neutral pH, T3 secretion apparatus should be assembled (Fig. 5 i). First, translocator proteins are secreted through T3SS but effector proteins are not, because regulatory complex blocks the effectors at the base of the needle (Fig. 5 ii). However once the translocator complex assembled a pore on the target cell membrane the T3SS can sense the neutral pH condition of the cytosol (Fig. 5 iii) and this pH elevation removes the regulatory complex from the base of the needle (Fig. 5 iv) to start secretion of effectors. After exposure, the cells were incubated at 37C in 5.0% CO2 and observed by fluorescent microscope.
Result and Discussion
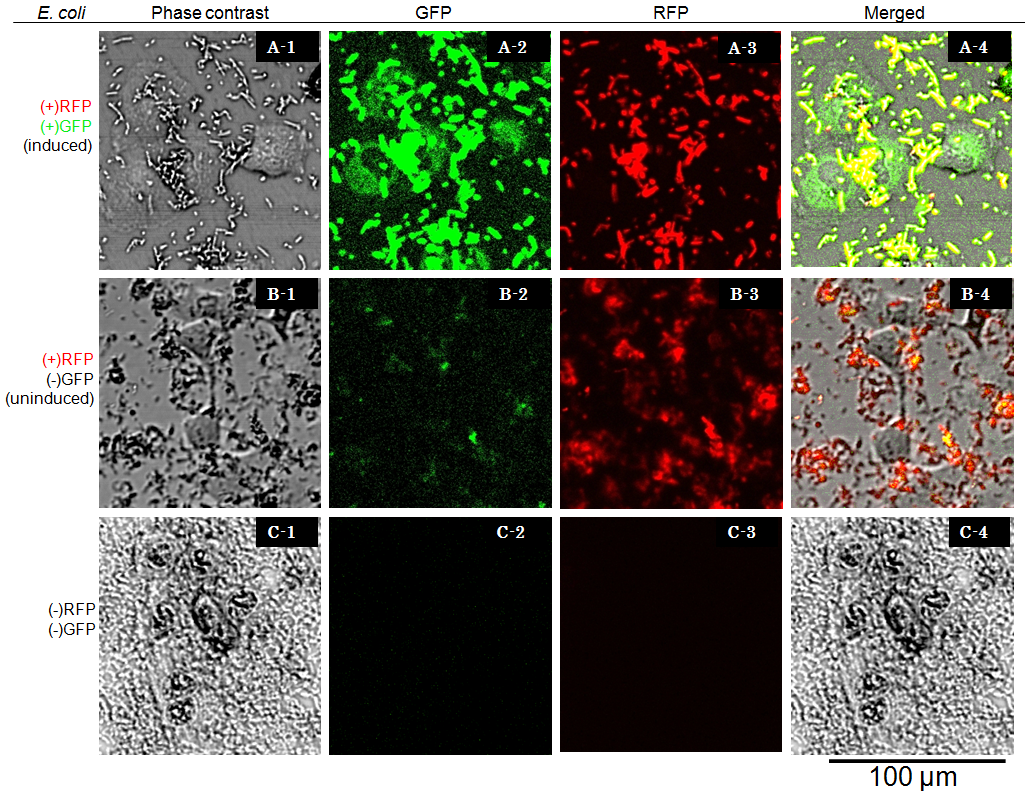
Cells were observed under Confocal Laser Scanning Microscope (OLYMPUS FV-1000D) with blue (473 nm) or green (559 nm) excitation lights at 7.5 hours after exposure. In the presence of arabinose E. coli expresses GFP and RFP, so bacterial cells are yellow( (Fig. 6 A-4), while GFP injected into target eukaryotic cells are green. Comparing Fig. 6 A-1 and A-4, GFP is located in the cytosol of target eukaryotic cells.
GFP translocated into the target cell didn’t show localization in the nucleus even 7.5 hours after exposure. These observations suggested that NLS domain in the GFP fusion construct we designed did not function as expected. This year we focus on this result, we aim to redesign the fusion protein vector, and to characterize its performance.
References
- Galán JE, Wolf-Watz H (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444: 567-573 Review [http://www.ncbi.nlm.nih.gov/pubmed/17136086 PubMed]
- Ghosh P (2004) Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev 68: 771-795 Review [http://www.ncbi.nlm.nih.gov/pubmed/15590783 PubMed]
- Miao EA, Miller SI (2000) A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc Natl Acad Sci U S A 97: 7539-7544 [http://www.ncbi.nlm.nih.gov/pubmed/10861017 PubMed]
- Rappl C, Deiwick J, Hensel M (2003) Acidic pH is required for the functional assembly of the type III secretion system encoded by Salmonella pathogenicity island 2. FEMS Microbiol Lett 226: 363-372 [http://www.ncbi.nlm.nih.gov/pubmed/14553934 PubMed]
- Waterman SR, Holden DW (2003) Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol 5: 501-511 Review [http://www.ncbi.nlm.nih.gov/pubmed/12864810 PubMed]
HokkaidoU_Japan iGEM 2010 on Wiki, Poster and Presentation.
 "
"