Team:HokkaidoU Japan/Project/RFC87
From 2011.igem.org
m (→Bsa I cloning site) |
|||
| (2 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div id="hokkaidou-right-content"> | <div id="hokkaidou-right-content"> | ||
| - | = | + | =Illustrations for the Bsa I cloning site= |
==Purpose== | ==Purpose== | ||
[[File:HokkaidoU_BsaI_Fig0.png|center]] | [[File:HokkaidoU_BsaI_Fig0.png|center]] | ||
| - | We designed "Bsa I cloning site" to accomplish following | + | We designed "Bsa I cloning site" to accomplish following requirements. |
| - | # | + | # Production a fusion protein |
| - | # | + | # Insertion must be universal |
| - | # | + | # Retain BioBrick properties of the construct |
| - | # | + | # Insertion of the parts only in correct direction |
| - | It is ideal that the insert is cut with BioBrick official restriction enzymes (requirement 2) however, cloning site must be cut without using those enzymes (requirement 3) | + | It is ideal that the insert is cut with BioBrick official restriction enzymes (requirement 2) however, cloning site must be cut without using those enzymes (requirement 3). Overhangs resulting from Bsa 1 digestion should not be the same to prevent ligation in incorrect direction(requirement 4). |
| - | |||
==Restriction Enzyme Bsa I== | ==Restriction Enzyme Bsa I== | ||
[[File:HokkaidoU_BsaI_Fig1.png|center]] | [[File:HokkaidoU_BsaI_Fig1.png|center]] | ||
| - | We came up with an idea of using restriction enzyme Bsa I. Bsa I recognition site and its cutting site are separated, and the sequence | + | We came up with an idea of using restriction enzyme Bsa I. Bsa I recognition site and its cutting site are separated, and the sequence on cutting site can be changed to suit different needs. |
| - | |||
==Design== | ==Design== | ||
[[File:HokkaidoU_BsaI_Fig2.png|center]] | [[File:HokkaidoU_BsaI_Fig2.png|center]] | ||
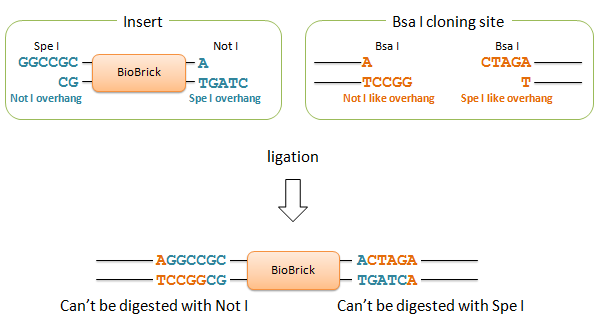
| - | We designed two Bsa I site to produce Not I like overhang and Spe I like overhang respectively. These cutting ends are ligated with Not I/ Spe I digested BioBricks in correct direction, and the product cannot be re-digested by Not I or Spe I. We can amplify every insert part using Prefix and Suffix primer set. | + | We designed two Bsa I site to produce Not I like overhang and Spe I like overhang respectively. These cutting ends are ligated with Not I/ Spe I overhangs of digested BioBricks in correct direction, and the product cannot be re-digested by Not I or Spe I. We can amplify every insert part using Prefix and Suffix primer set. |
| - | |||
==Problems== | ==Problems== | ||
[[File:HokkaidoU_BsaI_Fig3.png|center]] | [[File:HokkaidoU_BsaI_Fig3.png|center]] | ||
| Line 34: | Line 31: | ||
| - | |||
==Solution?== | ==Solution?== | ||
[[File:HokkaidoU_BsaI_Fig4.png|center]] | [[File:HokkaidoU_BsaI_Fig4.png|center]] | ||
| Line 40: | Line 36: | ||
| - | |||
==Solution!== | ==Solution!== | ||
[[File:HokkaidoU_BsaI_Fig5.png|center]] | [[File:HokkaidoU_BsaI_Fig5.png|center]] | ||
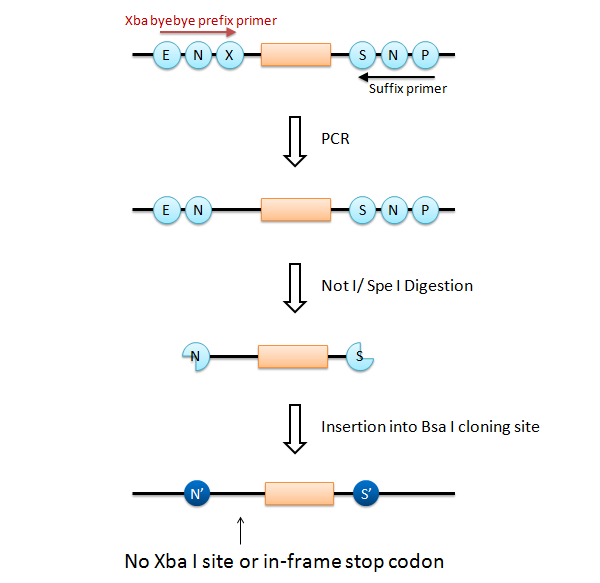
| - | We designed a new universal prefix primer named "Xba_byebye". This primer causes a point mutation to delete the Xba I site and its internal in-frame stop codon. Using the "Xba_byebye" prefix primer and the normal suffix primer, we can fulfill all the requirements. After PCR, the | + | We designed a new universal prefix primer named "Xba_byebye". This primer causes a point mutation to delete the Xba I site and its internal in-frame stop codon. Using the "Xba_byebye" prefix primer and the normal suffix primer, we can fulfill all the requirements. After PCR, the problematic template DNA can be removed by Dpn I digestion. |
| - | + | ==Application== | |
| - | == | + | |
[[File:HokkaidoU_BsaI_Fig6.png|center]] | [[File:HokkaidoU_BsaI_Fig6.png|center]] | ||
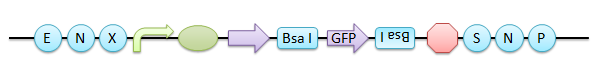
In this construct, the inserted GFP is interchangeable with another BioBrick part using Bsa I again, you can easily identify the replacement of the insert by the color of colonies. | In this construct, the inserted GFP is interchangeable with another BioBrick part using Bsa I again, you can easily identify the replacement of the insert by the color of colonies. | ||
| - | |||
We have submitted this method as [[Media:HokkaidoU_BBF_RFC_87.pdf|BBF RFC 87]]. | We have submitted this method as [[Media:HokkaidoU_BBF_RFC_87.pdf|BBF RFC 87]]. | ||
</div> | </div> | ||
{{Team:HokkaidoU_Japan/footer}} | {{Team:HokkaidoU_Japan/footer}} | ||
Latest revision as of 13:32, 15 December 2011
HokkaidoU Japan
iGEM 2011 Team of Hokkaido University
Contents |
- Abstract
- What`s T3SSDetailed information about T3SS and summary of our achievements on iGEM 2010
- Injection assay using onion cellsExperiments using plant cells are easier to perform than with mammalian ones
- Ready-to-inject backbone and Bsa I cloning siteReady-to-inject backbone and Bsa I cloning site enables easy fusion of T3S signal and protein
- GSK tag systemA neat injection assay using GSK tag, which can specifically detect successfully injected proteins
- Bsa I cloning site, RFC submissionDetailed documentation of costructing a BioBrick cloning site a BioBrick!
Illustrations for the Bsa I cloning site
Purpose
We designed "Bsa I cloning site" to accomplish following requirements.
- Production a fusion protein
- Insertion must be universal
- Retain BioBrick properties of the construct
- Insertion of the parts only in correct direction
It is ideal that the insert is cut with BioBrick official restriction enzymes (requirement 2) however, cloning site must be cut without using those enzymes (requirement 3). Overhangs resulting from Bsa 1 digestion should not be the same to prevent ligation in incorrect direction(requirement 4).
Restriction Enzyme Bsa I
We came up with an idea of using restriction enzyme Bsa I. Bsa I recognition site and its cutting site are separated, and the sequence on cutting site can be changed to suit different needs.
Design
We designed two Bsa I site to produce Not I like overhang and Spe I like overhang respectively. These cutting ends are ligated with Not I/ Spe I overhangs of digested BioBricks in correct direction, and the product cannot be re-digested by Not I or Spe I. We can amplify every insert part using Prefix and Suffix primer set.
Problems
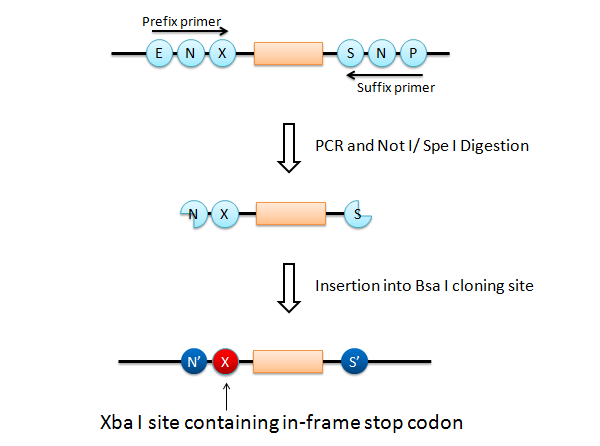
If this goes on, we have a serious problem that the final product has Xba I site after the Not I site (fail to fulfill the requirement 3). What is worse is that an in-frame stop codon appears on the Xba I site, just before the insert.
Solution?
To solve this problem, it is easy to add only a Not I site and a Spe I site at each end of the insert part. However, it requires different primer set that anneals each insert part (fail to fulfill the requirement 2).
Solution!
We designed a new universal prefix primer named "Xba_byebye". This primer causes a point mutation to delete the Xba I site and its internal in-frame stop codon. Using the "Xba_byebye" prefix primer and the normal suffix primer, we can fulfill all the requirements. After PCR, the problematic template DNA can be removed by Dpn I digestion.
Application
In this construct, the inserted GFP is interchangeable with another BioBrick part using Bsa I again, you can easily identify the replacement of the insert by the color of colonies.
We have submitted this method as BBF RFC 87.
 "
"