Team:Glasgow/Biofilm/Nissle
From 2011.igem.org
| (25 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Glasgow/Header}} | {{Team:Glasgow/Header}} | ||
<html> | <html> | ||
| - | <h1><i>E.coli</i> Nissle 1917</h1> | + | <h1><i>E. coli</i> Nissle 1917 - A novel biofilm-forming chassis</h1> |
| - | < | + | |
| - | < | + | <a name="top"></a><h6> Contents</h6> |
| - | < | + | <ul> <a href="#biofilms"><i>E.coli</i> Biofilms</a></ul> |
| - | < | + | <ul> <a href="#transform">Transforming Nissle</i></a></ul> |
| - | < | + | <ul> <a href="#registry">Nissle on the Registry</a></ul> |
| + | <ul> <a href="#ref2">References</a></ul> | ||
| + | |||
| + | <h6>Biofilm forming organisms</h6> | ||
| + | <ul> <a href="https://2011.igem.org/Team:Glasgow/Biofilm/P._aeruginosa">Pseudomonas aeruginosa</a></ul> | ||
| + | |||
| + | <h6> <a href="https://2011.igem.org/Team:Glasgow/BiofilmResults">Results obtained from experimentation</a></h6> | ||
| + | <h6><a href="https://2011.igem.org/Team:Glasgow/Biofilm">Back to Biofilms</a></h6> | ||
| + | <h6><a href="https://2011.igem.org/Team:Glasgow/Results">Back to Results</a></h6> | ||
| + | |||
| + | <table> | ||
<tr> | <tr> | ||
| - | <td> | + | <td><p> |
| - | < | + | When the team began thinking about our project in May, we decided that it would be interesting to work with biofilm forming bacteria, however, we encountered several problems before we were able to start measuring the effect of our biobricks.</p> |
| + | |||
| + | <p>Firstly, we needed a repeatable method through which to form biofilms. There are a large number of proposed methods available for growing biofilm. Unfortunately, many required specialised materials which we did not have access to or were prohibitively expensive. In the end, we developed a new protocol for growing biofilm based on a number of other protocols and the materials that we had available to us in the lab.</p> | ||
| + | |||
| + | <p>Next we required an assay through which to measure biofilm formation and dispersal. This required not only qualitative measurements (ie - has any biofilm formed at all?) but also in a manner which generates quantitative results. Again, other methods for this do already exist but were inaccessible to us because of cost or availability - because of this we generated a method for measuring formation and dispersal using the materials that we had available.</p> | ||
| + | |||
</td> | </td> | ||
<td> | <td> | ||
| - | <img src="http://farm7.static.flickr.com/ | + | <div><img src="http://farm7.static.flickr.com/6156/6170596909_c231821e8d_m.jpg" height="200px" width="250px" align="center" /><p><font size="1" color="grey"></div> |
| - | + | <div>Image 1: RFP Nissle cells plated in serial dilution</font></p></div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | <p><font size="1" color="grey"> | + | |
</td> | </td> | ||
| + | </tr> | ||
</table> | </table> | ||
| - | |||
| - | <p> | + | <p>When we came to actually attempting to form biofilm we encountered another problem: many of the lab strains of <i>E. coli</i> have selectively lost their ability to form biofilm due to chromosomal deletions. Originally <i>Pseudomonas aeruginosa</i> was considered as a chassis, but it was quickly discarded as a majority of the biobricks available in the registry have already been optimised for use in <i>E. coli</i>. Whilst researching different strains of <i>E. coli</i> with the ability to form biofilm a member of our team came across the Nissle 1917 strain (Hancock, 2010). It has previously been documented on multiple occasions to be non-pathogenic, safe for human consumption and is commercially available as Mutaflor tablets in Germany. Being an <i>E. coli</i> strain, is was fully expected to be suitable for transformation with the majority of standard biobricks</p> |
| + | <p>We obtained the cells from Gerhard Breves, Hanover Institute of Physiology, and succesfully extracted them from <a href="http://en.wikipedia.org/wiki/Mutaflor"><i>Mutaflor</i> Tablets</a>. These tablets are commercially available in Germany as a treatment for improving the gut lining, and <i>E. coli</i> have been shown to outcompete other biofilm-forming bacteria within the gut (Hancock, 2010) and to reduce the adhesion of Salmonella to porcine gut lining (Schierack et. al, 2011) </p> | ||
| + | <p>Mutaflor tablets are currently the only way to obtain Nissle cells! To extract the viable cells a single tablet was broken and the contents dissolved in 10ml of water. The resulting solution was then plated in serial dilution, ranging from undiluted to x 10<sup>-9</sup> on MacConkey's agar to select for the desired cells. It was found that a dilution between x 10<sup>-3</sup> and x 10<sup>-5</sup> yielded single colonies that we used in further experiments.</p> | ||
| + | <ul> <a href="#top">Return to top</a></ul> | ||
| + | <a name="biofilms"></a><h2><i>E. coli</i> Nissle 1917 Biofilms</h2> | ||
| + | <p>To prove biofilm formation glass slides were put into 50ml tubes containing 20ml of LB broth. The LB covered around a third of the glass slide, this is the area where the biofilm would form. The LB was then inoculated with 20μl of fresh overnight culture of <i>E. coli</i> Nissle. These tubes were then left on a bench top shaker at room temperature for 24 and 48 hours. The glass slides were then removed and the biofilm was stained by standard gram stain method. Photographs were taked of the stained biofilms.</p> | ||
| + | <p>Biofilm formation was also confirmed by SEM pictures that showed the extracellular matrix.</p> | ||
| + | </br> | ||
| + | <p>Once it had been shown that <i>E.coli</i> Nissle formed biofilms, a time series of biofilm formation was performed. The tubes containing the slides were left on the tabletop shaker for a set amount of time (time points ranged from 1hr to 24hrs). After the biofilm had grown its allotted time the glass slide was carefully removed and placed into a fresh 50ml tube with 25ml of LB (which completely covered the biofilm) and left to allow the bacteria to disperse for 1hr. The slide was then transferred to a fresh 50ml tube with 25ml of LB. The biofilm is scraped off using a spatula, and the resulting culture was vortexed for 1 minute to break up any residual chunks of biofilm. The culture containing the dispersed cells and the culture containing the broken-up biofilm were then plated in serial dilution to obtain an estimate of the number of cells in each culture.<p> Results showed that on average 48% of the cells within a biofilm will disperse once the biofilm is transferred into fresh media. The percentage dispersed was expected to be inversely correlated to the size of the biofilm, because a larger biofilm was expected to have a higher affinity for the cells it holds. However, the data shows that the percentage dispersed is relatively constant, and varies between 30-52% dispersal. | ||
| + | We are currently unsure whether we should be seeing a positive or negative correlation between the two.</p> | ||
| + | <p>The results are shown below.</p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
<div> | <div> | ||
| - | <img src="http://farm7.static.flickr.com/ | + | <table> |
| - | <p><font size="1" color="grey"> Graph | + | <tr> |
| - | </p> | + | <td> |
| - | <p>Chemically and Electrically competent <i>E. coli</i> Nissle cells were made and successfully transformed with RFP. Electroporation was notably more efficient than any chemical transformation. Problems were caused by the cells' tendency to clump together, which we assume is caused by their long fimbriae that were visible on the SEM pictures. | + | <img src="http://farm7.static.flickr.com/6175/6171301852_67b79bf53b.jpg"> |
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><img src="http://farm7.static.flickr.com/6160/6171301936_2ba997846b_b.jpg" alt="Graph of percentage dispersal from biofilm" width="850" height="500"> | ||
| + | <p><font size="1" font color="grey"> Graph 1: The total percentage of cells dispersed when a biofilm is transferred into new media vs time in hours </font></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <ul> <a href="#top">Return to top</a></ul> | ||
| + | <a name="transform"></a><h2>Transforming Nissle</h2> | ||
| + | <p><p>Chemically and Electrically competent <i>E. coli</i> Nissle cells were made and successfully transformed with <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_J04450">RFP (BBa_J04450)</a>. Electroporation was notably more efficient than any chemical transformation. Problems were caused by the cells' tendency to clump together, which we assume is caused by their long fimbriae that were visible on the SEM pictures. | ||
this problem can be countered by either vortexing or sonicating the culture before plating them. We expect sonication to be the more efficient method.</p> | this problem can be countered by either vortexing or sonicating the culture before plating them. We expect sonication to be the more efficient method.</p> | ||
| - | <p>The transformed <i>E.coli</i> Nissle cells showed expression of RFP, in culture and within a biofilm, proving that they can express standard <i>E.coli</i> constructs. The results can be seen in | + | <p>The transformed <i>E.coli</i> Nissle cells showed expression of RFP, in culture and within a biofilm, proving that they can express standard <i>E.coli</i> constructs. The results can be seen in Images 2 through 4 below.</p> |
<div> | <div> | ||
<table align="centre"> | <table align="centre"> | ||
| Line 42: | Line 73: | ||
<td> | <td> | ||
<img src="http://farm7.static.flickr.com/6161/6168749805_72b6068728_m.jpg"/> | <img src="http://farm7.static.flickr.com/6161/6168749805_72b6068728_m.jpg"/> | ||
| - | <p><font size="1" color="grey"> | + | <p><font size="1" color="grey">Image 2: RFP transformed <i>E.coli</i> Nissle </br>overnight culture </font></p> |
</td> | </td> | ||
<td> | <td> | ||
<img src="http://farm7.static.flickr.com/6179/6167447454_14246d184b_m.jpg"/> | <img src="http://farm7.static.flickr.com/6179/6167447454_14246d184b_m.jpg"/> | ||
| - | <p><font size="1" color="grey"> | + | <p><font size="1" color="grey"> Image 3: A 24-hour biofilm of RFP </br><i>E.coli</i> Nissle </font></p> |
</td> | </td> | ||
<td> | <td> | ||
<img src="http://farm7.static.flickr.com/6180/6168749811_6f05839649_m.jpg"/> | <img src="http://farm7.static.flickr.com/6180/6168749811_6f05839649_m.jpg"/> | ||
| - | <p><font size="1" color="grey"> | + | <p><font size="1" color="grey"> Image 4: Streaked RFP transformed </br><i>E.coli</i> Nissle </font></p> |
</td> | </td> | ||
</table> | </table> | ||
</div> | </div> | ||
| - | |||
| - | |||
<p> Unfortunately we were unable to transform the Nissle cells with our novel biobricks due to time constraints, and were therefore unable to operate our synthetic system within the biofilm.</p> | <p> Unfortunately we were unable to transform the Nissle cells with our novel biobricks due to time constraints, and were therefore unable to operate our synthetic system within the biofilm.</p> | ||
| - | <h4>Reference</h4> | + | </br> |
| + | <ul> <a href="#top">Return to top</a></ul> | ||
| + | <a name="registry"></a><h2>Nissle on the Registry</h2> | ||
| + | <p> Since Nissle have proven to be transformable, and have distinct biofilm-forming abilities we have made the cells available to the Registry as a novel chassis for use in further research and projects involving biofilms.</p> | ||
| + | <b><p> Continue to<a href="https://2011.igem.org/Team:Glasgow/BiofilmResults"> Summary and Results </a></b></p> | ||
| + | |||
| + | <a name="ref2"></a><h4>Reference</h4> | ||
<p>Hancock, V., Dahl, M. & Klemm, P., 2010. Probiotic <i>Escherichia coli</i> strain Nissle 1917 outcompetes intestinal pathogens during biofilm formation.Journal of Medical Microbiology. Vol 59, Jan 2010 pp 392-399. </p> | <p>Hancock, V., Dahl, M. & Klemm, P., 2010. Probiotic <i>Escherichia coli</i> strain Nissle 1917 outcompetes intestinal pathogens during biofilm formation.Journal of Medical Microbiology. Vol 59, Jan 2010 pp 392-399. </p> | ||
| + | <p>Schierack P, Kleta S, Tedin K, Babila JT, Oswald S, Oelschlaeger TA, Hiemann R, Paetzold S, Wieler LH. 2011.<a href="http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0014712"> E. coli Nissle 1917 Affects Salmonella adhesion to porcine intestinal epithelial cells.</a> | ||
| + | |||
| + | <ul> <a href="#top">Return to top</a></ul> | ||
| + | |||
</html> | </html> | ||
Latest revision as of 03:08, 22 September 2011

E. coli Nissle 1917 - A novel biofilm-forming chassis
Contents
Biofilm forming organisms
Results obtained from experimentation
Back to Biofilms
Back to Results
When the team began thinking about our project in May, we decided that it would be interesting to work with biofilm forming bacteria, however, we encountered several problems before we were able to start measuring the effect of our biobricks. Firstly, we needed a repeatable method through which to form biofilms. There are a large number of proposed methods available for growing biofilm. Unfortunately, many required specialised materials which we did not have access to or were prohibitively expensive. In the end, we developed a new protocol for growing biofilm based on a number of other protocols and the materials that we had available to us in the lab. Next we required an assay through which to measure biofilm formation and dispersal. This required not only qualitative measurements (ie - has any biofilm formed at all?) but also in a manner which generates quantitative results. Again, other methods for this do already exist but were inaccessible to us because of cost or availability - because of this we generated a method for measuring formation and dispersal using the materials that we had available. |
 Image 1: RFP Nissle cells plated in serial dilution
|
When we came to actually attempting to form biofilm we encountered another problem: many of the lab strains of E. coli have selectively lost their ability to form biofilm due to chromosomal deletions. Originally Pseudomonas aeruginosa was considered as a chassis, but it was quickly discarded as a majority of the biobricks available in the registry have already been optimised for use in E. coli. Whilst researching different strains of E. coli with the ability to form biofilm a member of our team came across the Nissle 1917 strain (Hancock, 2010). It has previously been documented on multiple occasions to be non-pathogenic, safe for human consumption and is commercially available as Mutaflor tablets in Germany. Being an E. coli strain, is was fully expected to be suitable for transformation with the majority of standard biobricks
We obtained the cells from Gerhard Breves, Hanover Institute of Physiology, and succesfully extracted them from Mutaflor Tablets. These tablets are commercially available in Germany as a treatment for improving the gut lining, and E. coli have been shown to outcompete other biofilm-forming bacteria within the gut (Hancock, 2010) and to reduce the adhesion of Salmonella to porcine gut lining (Schierack et. al, 2011)
Mutaflor tablets are currently the only way to obtain Nissle cells! To extract the viable cells a single tablet was broken and the contents dissolved in 10ml of water. The resulting solution was then plated in serial dilution, ranging from undiluted to x 10-9 on MacConkey's agar to select for the desired cells. It was found that a dilution between x 10-3 and x 10-5 yielded single colonies that we used in further experiments.
E. coli Nissle 1917 Biofilms
To prove biofilm formation glass slides were put into 50ml tubes containing 20ml of LB broth. The LB covered around a third of the glass slide, this is the area where the biofilm would form. The LB was then inoculated with 20μl of fresh overnight culture of E. coli Nissle. These tubes were then left on a bench top shaker at room temperature for 24 and 48 hours. The glass slides were then removed and the biofilm was stained by standard gram stain method. Photographs were taked of the stained biofilms.
Biofilm formation was also confirmed by SEM pictures that showed the extracellular matrix.
Once it had been shown that E.coli Nissle formed biofilms, a time series of biofilm formation was performed. The tubes containing the slides were left on the tabletop shaker for a set amount of time (time points ranged from 1hr to 24hrs). After the biofilm had grown its allotted time the glass slide was carefully removed and placed into a fresh 50ml tube with 25ml of LB (which completely covered the biofilm) and left to allow the bacteria to disperse for 1hr. The slide was then transferred to a fresh 50ml tube with 25ml of LB. The biofilm is scraped off using a spatula, and the resulting culture was vortexed for 1 minute to break up any residual chunks of biofilm. The culture containing the dispersed cells and the culture containing the broken-up biofilm were then plated in serial dilution to obtain an estimate of the number of cells in each culture.
Results showed that on average 48% of the cells within a biofilm will disperse once the biofilm is transferred into fresh media. The percentage dispersed was expected to be inversely correlated to the size of the biofilm, because a larger biofilm was expected to have a higher affinity for the cells it holds. However, the data shows that the percentage dispersed is relatively constant, and varies between 30-52% dispersal. We are currently unsure whether we should be seeing a positive or negative correlation between the two.
The results are shown below.
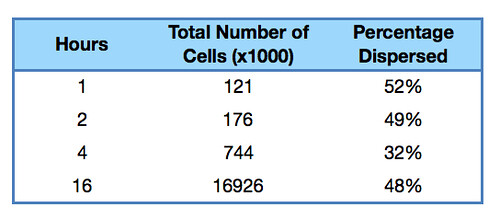
|
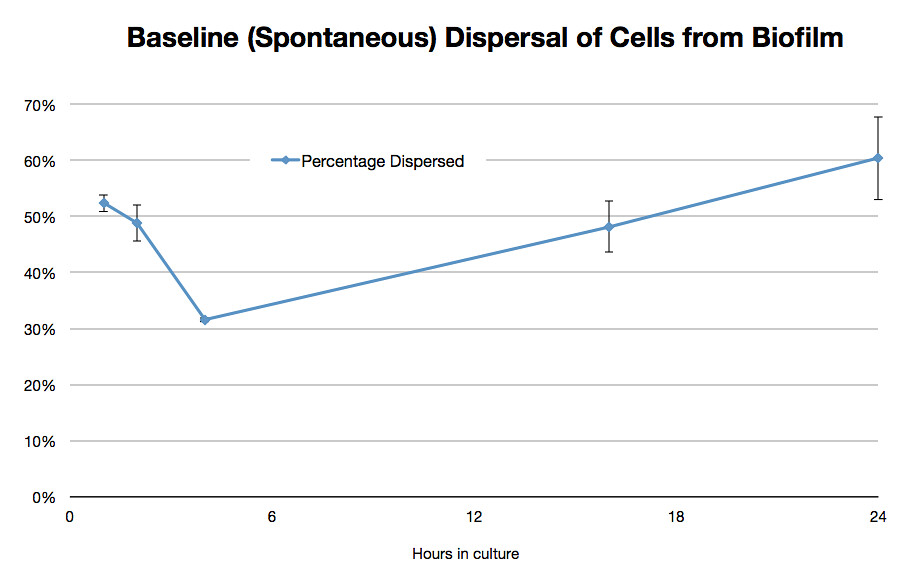
Graph 1: The total percentage of cells dispersed when a biofilm is transferred into new media vs time in hours |
Transforming Nissle
Chemically and Electrically competent E. coli Nissle cells were made and successfully transformed with RFP (BBa_J04450). Electroporation was notably more efficient than any chemical transformation. Problems were caused by the cells' tendency to clump together, which we assume is caused by their long fimbriae that were visible on the SEM pictures. this problem can be countered by either vortexing or sonicating the culture before plating them. We expect sonication to be the more efficient method.
The transformed E.coli Nissle cells showed expression of RFP, in culture and within a biofilm, proving that they can express standard E.coli constructs. The results can be seen in Images 2 through 4 below.

Image 2: RFP transformed E.coli Nissle overnight culture |
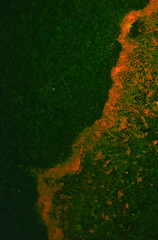
Image 3: A 24-hour biofilm of RFP E.coli Nissle |
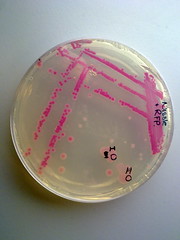
Image 4: Streaked RFP transformed E.coli Nissle |
Unfortunately we were unable to transform the Nissle cells with our novel biobricks due to time constraints, and were therefore unable to operate our synthetic system within the biofilm.
Nissle on the Registry
Since Nissle have proven to be transformable, and have distinct biofilm-forming abilities we have made the cells available to the Registry as a novel chassis for use in further research and projects involving biofilms.
Continue to Summary and Results
Reference
Hancock, V., Dahl, M. & Klemm, P., 2010. Probiotic Escherichia coli strain Nissle 1917 outcompetes intestinal pathogens during biofilm formation.Journal of Medical Microbiology. Vol 59, Jan 2010 pp 392-399.
Schierack P, Kleta S, Tedin K, Babila JT, Oswald S, Oelschlaeger TA, Hiemann R, Paetzold S, Wieler LH. 2011. E. coli Nissle 1917 Affects Salmonella adhesion to porcine intestinal epithelial cells.
 "
"
